Fig. 1.
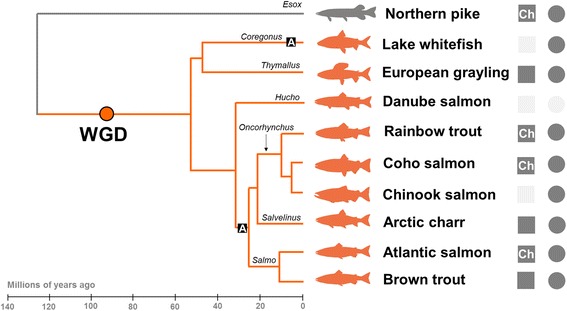
The comparative-evolutionary framework of FAASG. Shown are the initial target species for functional annotation (see Table 1) and their evolutionary relationships (time-calibrated tree after [7]). The selected species come from all three salmonid subfamilies. The position of the salmonid-specific WGD is highlighted (after [7, 9, 10]), along with Latin names of genera. Additional salmonid species that are future potential targets for functional annotation are not shown. Two lineages where anadromous life-history is thought to have evolved independently are highlighted ‘A’ (after [47]). The status of genomics resources are shown to the right of the tree: squares and circles indicate genome and transcriptome assemblies, respectively (dark grey = resource either published or close to being published; light grey = resource under active development; ‘Ch’ = chromosome-anchored genome assembly)
