Extended Data Figure 1. Dnd1 expression, conservation and cell lines.
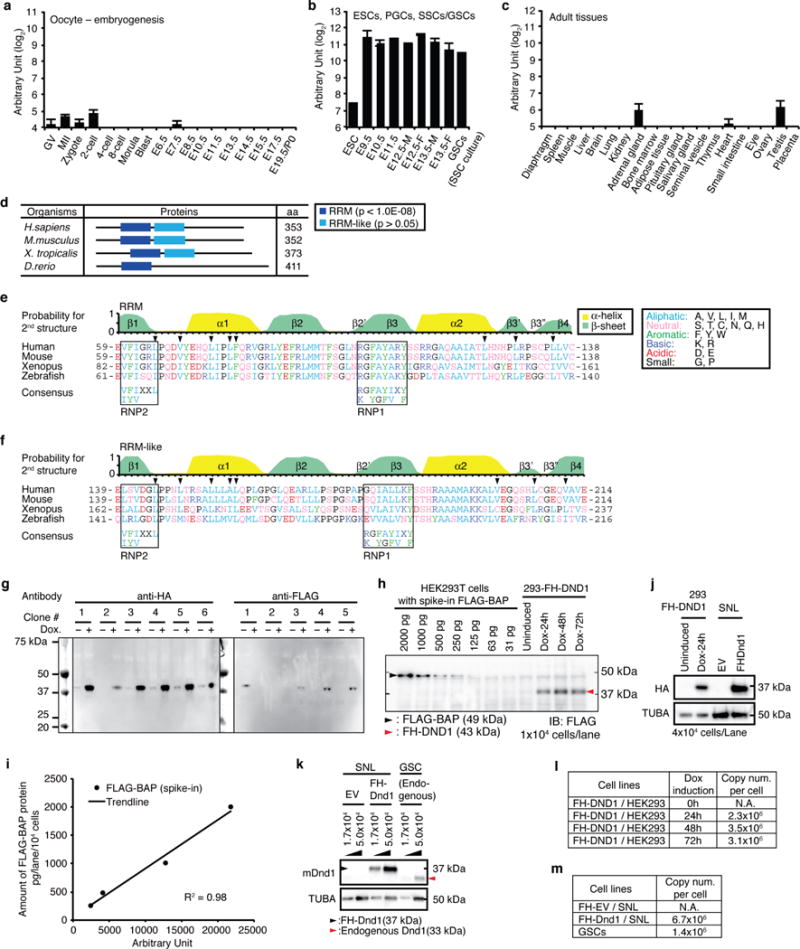
a–c, Dnd1 mRNA abundance determined by RNA-seq, a, during embryogenesis from oocytes to birth (Postnatal day 0, P0) at embryonic day (E) 19.5, b, in embryonic stem cells (ESCs), developing PGCs isolated from E9.5 to E13.5 embryos, and in vitro culture of spermatogonial stem cells (germline stem cells, GSCs), and c, in adult tissues. Error bars indicate SEM (3–5 replicates). Abbreviations: GV, germinal vesicle oocytes; MII, mature metaphase II oocytes; -M, male; -F, female. d, Domain organization of DND1 proteins from representative vertebrates determined by SMART (Normal mode, http://smart.embl.de/)51. The peptide length in aa is indicated. RRM (p <10−8) and RRM-like (p >0.05) domains are represented as blue and light blue boxes, respectively. e,f, Structure and amino acid sequence comparison of DND1 RRM (e) and RRM-like (f) domains between human and other vertebrates. The top panel indicates the probability for α-helix (yellow) and β-sheet (green) formation of human DND1 predicted by NetSurfP52 (http://www.cbs.dtu.dk/services/NetSurfP/), as well as information on secondary structure elements required to form the RRM-fold53–55. The arrowheads indicate the residues forming the hydrophobic interactions for the β3′-β3″ β-hairpin. Similarity in the amino acid composition was assessed based on the classification of the amino acid residues as shown in the legend; aliphatic (A and V), aromatic (F, Y, and W), basic (K and R), acidic (D and E), neutral (S, T, C, N, Q, and H), or small (G and P). The highly conserved ribonucleoprotein motifs, RNP2 and RNP1, involved in protein-RNA interaction, are indicated in black boxes, together with their consensus sequences, (L/I)-(Y/F)-(V/I)-X-X-L and (R/K)-G-(F/Y)-(G/A)-(F/Y)-(I/V)-X-(F/Y), respectively53,54. Note that aromatic residues in RNP1 and 2 and basic residues in RNP1 are not conserved in the RRM-like domain (f). Additionally, compared with the RRM domain, the probability to form β3′ and β3″ sheets in the RRM-like domain is low and the aliphatic residues (V200 and L206 in human) constraining the β3′-β3″ β-hairpin (arrowheads) are less conserved. g, Screen for stable HEK293 cell clones inducibly expressing FLAG-HA-tagged (FH)-DND1. Clone 4 was selected for PAR-CLIP experiments and clones 1 and 4 were used for apoptosis assays. h, The indicated amounts of purified FLAG-tagged protein (FLAG-BAP) were spiked into lysates from parental Flp-In HEK293-TRex cells. Expression of FH-DND1 in stable HEK293 cell lines was induced by doxycycline for the indicated amount of time. The equivalent of 104 cells was loaded into each well of the SDS-PAGE for immunoblotting. FLAG-BAP and FH-DND1 were detected using anti-FLAG M2 antibodies. i, Calibration curve from the signal intensity of FLAG-BAP (h) quantified by ImageJ software and plotted against the amount of FLAG-BAP (pg/104 cells). The regression line and coefficient of determination (R2) are shown. j, Protein expression of human or mouse FH-DND1 in HEK293 cells with or without Dox treatment for the indicated time or in mouse SNL cells, respectively. 4×104 cells were loaded in each lane of the SDS-polyacrylamide gel. Immunoblot from was probed with anti-HA antibodies (upper panel). The same membrane was re-probed for α-tubulin (TUBA) as loading control. k, Protein expression of mouse FH-DND1 (black arrowhead) in SNL cells or endogenous mouse DND1 (red arrowhead) in GSCs, respectively. SNL cells transformed with empty vector (EV) was used as negative control. Immunoblot was probed with anti-DND1-antibody9. TUBA served as loading control. Lysate from the indicated amount of cells was loaded onto each lane of the SDS-polyacrylamide gel. l, Copy numbers per cell of FH-DND1 in HEK293 cells after induction of transgene expression for the indicated amount of time. Values were determined by quantification (h,j). m, Copy numbers per cell of FH-DND1 and endogenous DND1 in mouse cells, determined by quantification of the DND1 band from (k).
