Extended Data Figure 6. Analysis of DND1 function in adult murine spermatogonia.
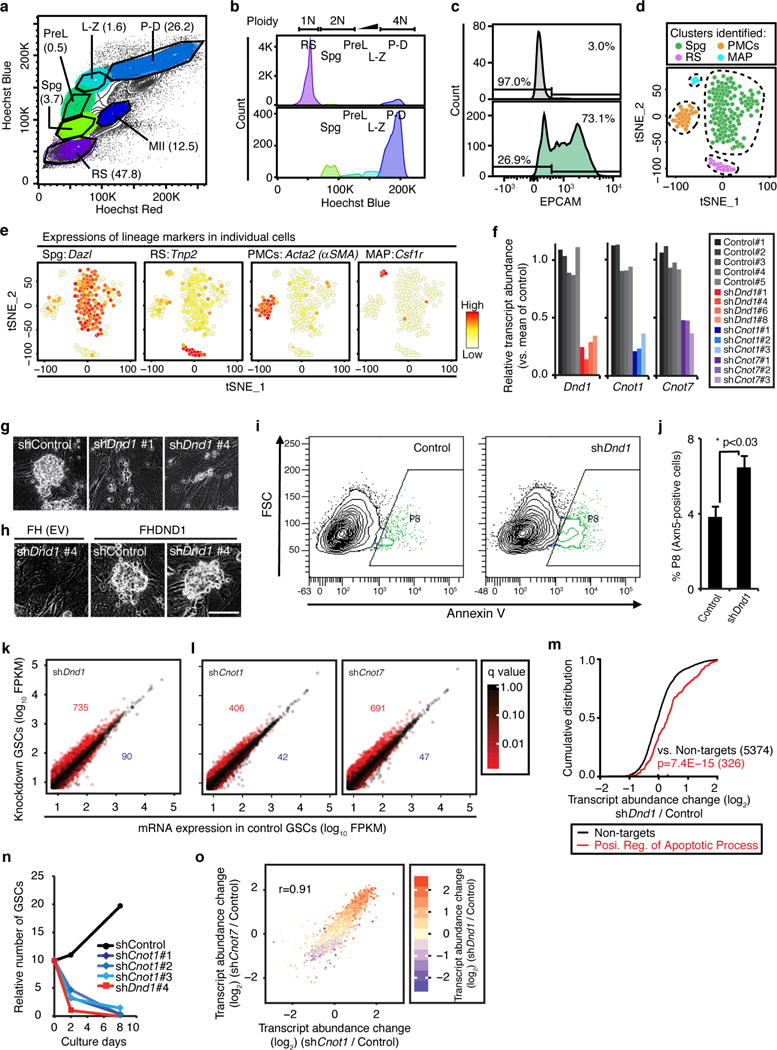
a, Hoechst profile of seminiferous tubular cells visualized in a “Hoechst Blue”/“Hoechst Red” contour plot. Numbers in parentheses indicate percentage of each population. Spermatogenic cells show heterogeneous morphologies, intracellular complexities, and DNA contents reflecting meiotic cell cycle. Spg, spermatogonia; PreL, preleptotene spermatocytes; L-Z, leptotene-zygotene spermatocytes; P-D, pachytene-diplotene spermatocytes; MII, meiosis II spermatocytes; RS, round spermatids. Further analysis of the Spg fraction is presented in Fig. 3. b, DNA content of the gated spermatogenic cells in (a) reveals successful separation of developing spermatogenic cells. c, Histogram of EPCAM+ spermatogonia in Spg population is shown in lower panel. The upper panel represents unstained control from the same cell suspension. d, t-SNE analysis of single-cell RNA-seq experiments from sorted spermatogonia (Spg from (a)) reveals specific clusters representing the indicated cell types. Abbreviations: Spg, spermatogonia; RS, round spermatids; PMCs, peritubular myoid cells; MAPs, macrophages. e, Heatmap representation of the expression of indicated genes on the tSNE plot. Cell types from (c) were identified according to the expression levels of marker genes. In total, 187 cells were analysed, and 115 cells of these cells (61%) were determined as actual spermatogonia according to expressions of lineage marker genes including Dazl. Numbers of other cell populations were: RS, 16 cells; PMCs, 29 cells; MAP, 5 cells. f, Knockdown efficiency of four different Dnd1-shRNA constructs (red) and three different Cnot1- and Cnot7-shRNA constructs (blue and purple, respectively) compared to five control shRNAs (grey) assessed by RNA-seq. g, Appearance of GSCs four days after transduction with the indicated shRNA (bar, 50 μm). h, Appearance of GSCs transduced with the indicated shRNA, as well as with an empty (upper panel) and FH-Dnd1 cDNA construct (lower panel; bar, 50 μm). i, Representative FACS contour plots for shRNA-transduced GSCs with the signal intensity of Annexin V on the x-axis (log10 scale) and the forward scatter (FSC) on the y-axis. Annexin V-positive cells represent early apoptotic cells and are gated as P8 population (green). j, The mean % population in P8 in three replicates of shDnd1 and Control are shown with SEMs. p<0.03 (Student’s t-test). k, Scatterplot comparing the average FPKM value of GSCs treated with shDnd1 and control shRNAs. The numbers of two-fold up- or down-regulated genes with p <0.05 are indicated in red or blue, respectively. l, Scatterplots comparing the mean FPKM value of (left panel) shCnot1 (three replicates) and (right panel) shCnot7 (three replicates) with control (five replicates). Transcripts are coloured according to q-value of transcript abundance change. m, CDF of transcript abundance changes between GSCs treated with shDnd1 and control shRNAs. Black line represents transcripts not targeted by DND1 and the red line represents targets classified as positive regulators of apoptosis according to GO (GO:0043065). The median transcript abundance change is indicated by a dot on the x-axis. p-values were determined by the MWU test. n, Proliferation of shRNA-transduced GSCs. The cumulative numbers of shControl-, shDnd1, or shCnot1-transduced GSCs were shown in black, red, or blue coloured dots, respectively. shRNAs were transduced by using lentiviral (LV) vectors at d0. 25 ng/ml puromycin was added at d2 to the culture medium to select stable transformants. o, Scatterplot similar to Fig. 3g–h of the log2 of transcript abundance changes of shCnot1/Control and shCnot7/Control. Pearson’s correlation coefficient is indicated. Transcripts are coloured according to the log2 of transcript abundance change of shDnd1/Control.
