Extended Data Figure 7. DND1 target mRNAs are silenced in developing PGC and Cas9-mediated gene knockout of Dnd1 in murine ESCs.
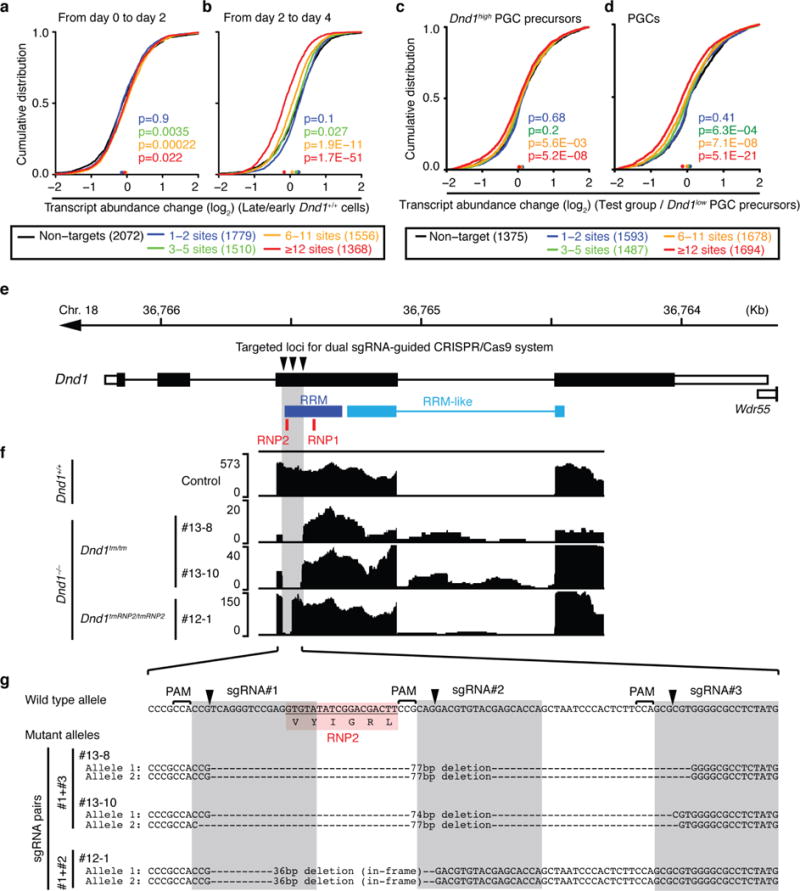
a–b, mRNA-expression change between (a) d0 and d2 PGCLC and (b) d2 and d4 PGCLC induced from EpiLCs was determined by RNA-seq (Supplementary Table 6). Expressed mouse DND1 PAR-CLIP target mRNAs (FPKM>5) were binned according to the number of binding sites (colored lines) and compared to expressed non-targets (FPKM>5, black line). The empirical cumulative distribution function (CDF) was plotted. The median transcript abundance change is indicated by a dot on the x-axis. p-values were determined by the MWU test. c, mRNA abundance change between Dnd1high- and Dnd1low-PGC-precurcors at E6.5–6.75 (n=6 each) was determined previously by single cell microarray analysis (Supplementary Tables 7 and 9). Expressed mouse DND1 PAR-CLIP target mRNAs (log2 array signal intensity >8) were binned and compared to expressed non-targets (log2 array signal intensity >8, black line) and the cumulative distribution function plotted as described in (a). d, Same as (c), except mRNA abundance change in specified PGCs at E7.25 (n=8) compared to Dnd1low-PGC precursor cells (n=6) was shown. e, Genome browser track showing the gene structure of Dnd1 on mouse chromosome 18. The bottom track indicates the regions encoding the RNA binding domain of DND1. RNP1 and RNP2 motifs contacting RNA (Extended Data Fig. 2). f, RNA-seq tracks for the same region as (a) showing the deletions induced by the sgRNAs for three different clones. g, Sanger DNA sequencing of the indicated region confirms successful deletions by the sgRNAs predicting to cut at the indicated positions. #13-8 and -10 showed targeted deletion of RNP2 associated with frame shift, resulting in the generation of null mutations in both alleles (Dnd1tm/tm). #12-1 has in-frame deletions for loss-of-RNP2 (Dnd1tmRNP2/tmRNP2). There was no distinguishable phenotype between these mutant mice in the germline development (Extended Data Fig. 8 and data not shown) and thus, we combined data from these three lines referred to as Dnd1−/−.
