Figure 4. MEcPP and SA alter jasmonate biosynthesis and signaling.
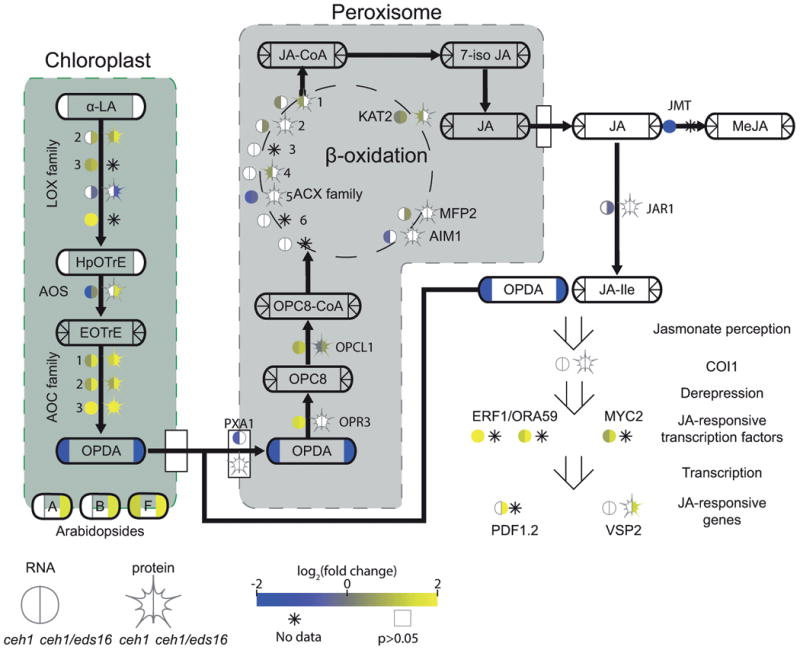
A pathway schematic for JA biosynthesis and breakdown, with transcription factors/enzymes shown by bare text with a circle, representing RNA, and star, representing protein levels. Metabolites are shown by rounded rectangles. For metabolites, RNA, and protein, the log2-fold change for ceh1 vs parent (left) and ceh1/eds16 vs. parent (right) is represented by shading, from -2 (darkest) to 2 (lightest). Non-detectable products in each section are shown by an asterisk, while empty symbols with grey borders represent non-significant fold changes relative to parent. Arrows represent chemical conversions, catalyzed by the enzymes shown on the line, or metabolite transport. Peroxisome-localized steps in conversion of 12-OPDA to JA are shown in the grey box, while simplified chloroplast-localized steps leading to 12-OPDA synthesis are in the green. Open arrows represent metabolite/protein, protein/protein, and protein/DNA (transcription regulation) interactions. Abbreviations: α-LA: α-linolenic acid, LOX: lipoxygenase, HpOTrE: hydroperoxyoctadecatrienoic acid, AOS: allene oxide synthase, EOTrE: epoxyoctadecatrienoic acid, AOC: allene oxide cyclase, OPDA: oxophytodienoic acid, PXA1: peroxisomal ABC transporter 1, OPR3: OPDA reductase 1, OPC8: oxopentenylcyclopentenyloctanoate, OPCL1: OPC8 coA ligase1, ACX: acyl-coA oxidase, KAT2: ketoacyl-coA thiolase 2, MFP2: multifunctional protein 2, AIM1: abnormal inflorescence meristem 1, MeJA: methyl-JA, COI1: coronatine insensitive 1, ERF1: ethylene response factor 1, ORA59: octadecanoid-responsive Arabidopsis AP2/ERF 59, MYC2: (=JIN1) MYC-domain containing transcription factor, PDF1.2: plant defensing 1.2, VSP2: vegetative storage protein 2.
