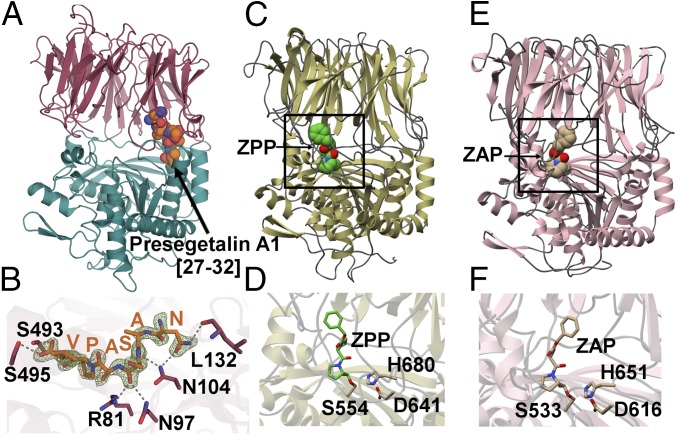
Fig. 3.
(A) The 1.9-Å PCY1 structure with the α/β hydrolase domain shown in green and β-propeller domain shown in red. (B) The C-terminal six amino acids of the presegetalin A1 [14–32] substrate (orange) is bound near the hinge region of PCY1. A simulated annealing difference Fourier maps (Fo-Fc), calculated with the coordinates of the peptide omitted, is superimposed and contoured to 2.5σ (blue). (C) Structure of the canonical POP enzyme from S. scorfa (PDB ID code 1QFS) (38, 39) bound to covalent inhibitor ZPP, with the active site demarcated in the rectangle, and (D) a close-up view of the active site. (E) Structure of the bacterial enzyme from M. xanthus (PDB ID code 2BKL) (40) bound to covalent inhibitor Z-Ala-prolinal, with the active site demarcated, and (F) a close-up view of the active site.