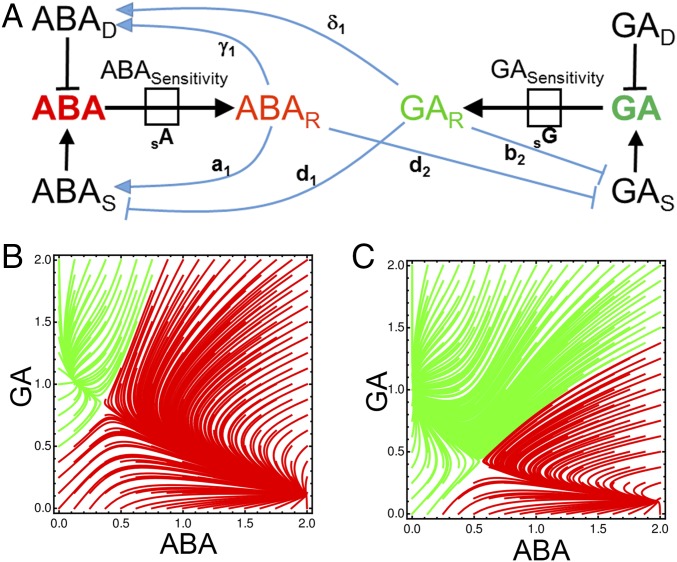
Fig. 1.
Mathematical model of hormone metabolic interactions underlying fate switching in Arabidopsis seeds. (A) Schematic outlining the relationships between the components of the hormone metabolic model. The hormone ABA is shown in red and bold, and the hormone GA is shown in green and bold. The response to each of these hormones, each comprising expression of several genes, is denoted by the subscript R (ABAR and GAR). The degradation of each hormone is indicated by the subscript D (ABAD and GAD), synthesis is indicated by the subscript S (ABAS and GAS), and sensitivity is written in full (ABASensitivity and GASensitivity). The directions of the arrows in the model are defined using microarray data describing ABA and GA application to Arabidopsis seeds (17, 47) and the associated gene expression changes for components representing each component (SI Appendix, Supplementary Fig. 1). GA degrading gene expression (GA 2-oxidase) was not detected at significant levels in germinating or dormant seeds. Attractor basins describing the dynamics of metabolic poise under a dynamic model parameterized by observations for deeply dormant seeds (B) and less dormant seeds (C) are shown. Axes indicate ABA and GA hormone abundance, and trajectories give the dynamics of the system starting from a given point and converging on one of two stable states.