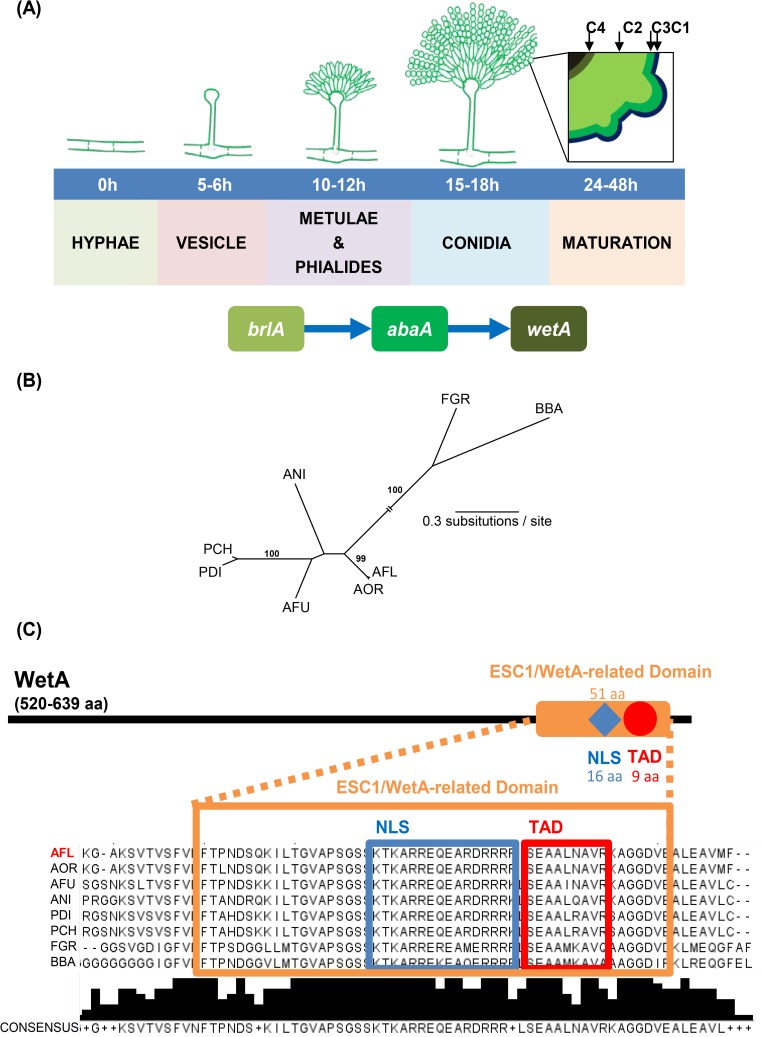
Fig 1. WetA is required for proper conidial maturation and contains both a transcription activation domain and a nuclear localization signal in a variety of fungi.
(A) A model for the roles of the central regulators in Aspergillus conidiogenesis. WetA is activated by AbaA and is responsible for conidia wall maturation. The black square illustrates the graphic view of the wall structure of the mature conidium, including the crenulated electron-dense outer layer C1, the carbohydrate-condensed layer C3, the electron-thin layer C2, and the innermost layer C4. Note: some Aspergillus species lack metulae (ex. A. parasiticus), and some species can have both metulae-phialides or phialides-only conidiophores (ex. some A. flavus variants) [11]. (B) Unrooted phylogeny of WetA amino acid sequences of A. flavus NRRL3357 XP_002383329.1 (AFL), A. fumigatus Af293 XP_751508.1 (AFU), A. nidulans FGSC4 XP_659541.1 (ANI), A. oryzae RIP40 XP_001816745.1 (AOR), Penicillium chrysogenum Wisconsin 54–1255 XP_002564365.1 (PCH), P. digitatum Pd1 XP_014534725.1 (PDI), Fusarium graminearum PH-1 I1S0E2.2 (FGR), and Beauveria bassiana ARSEF 2860 XP_008599445.1 (BBA) [12–18]. The sequences were aligned using MAFFT, version 7.1.5 [19]. The WetA protein phylogeny was calculated using the maximum likelihood optimality criterion, as implemented in PAUP [20], version 4.0a152; we used the WAG model of amino acid evolution [21], with empirical amino acid frequencies and allowing for rate heterogeneity among sites. Values near internal branches correspond to bootstrap support values (only values above 70% are shown). Branch lengths correspond to the estimated number of amino acid substitutions per site–the internal branch leading to the FGR and BBA sequences has been truncated for optimal visualization. (C) The predicted WetA protein architecture. The red circle and the red box represent the transcription activation domain (TAD) which was predicted by 9aaTAD using the “Less stringent Pattern” setting [22]. The blue diamond and the blue box represent the nuclear localization signal (NLS) predicted by NLStradamus using the 4 state HMM static model [23]. The orange rectangle and the orange box represent the ESC1/WetA-related domain (PTHR22934) predicted by the PANTHER classification system [24]. The consensus sequence and the consensus histogram are shown under the amino acid sequence multiple sequence alignment.