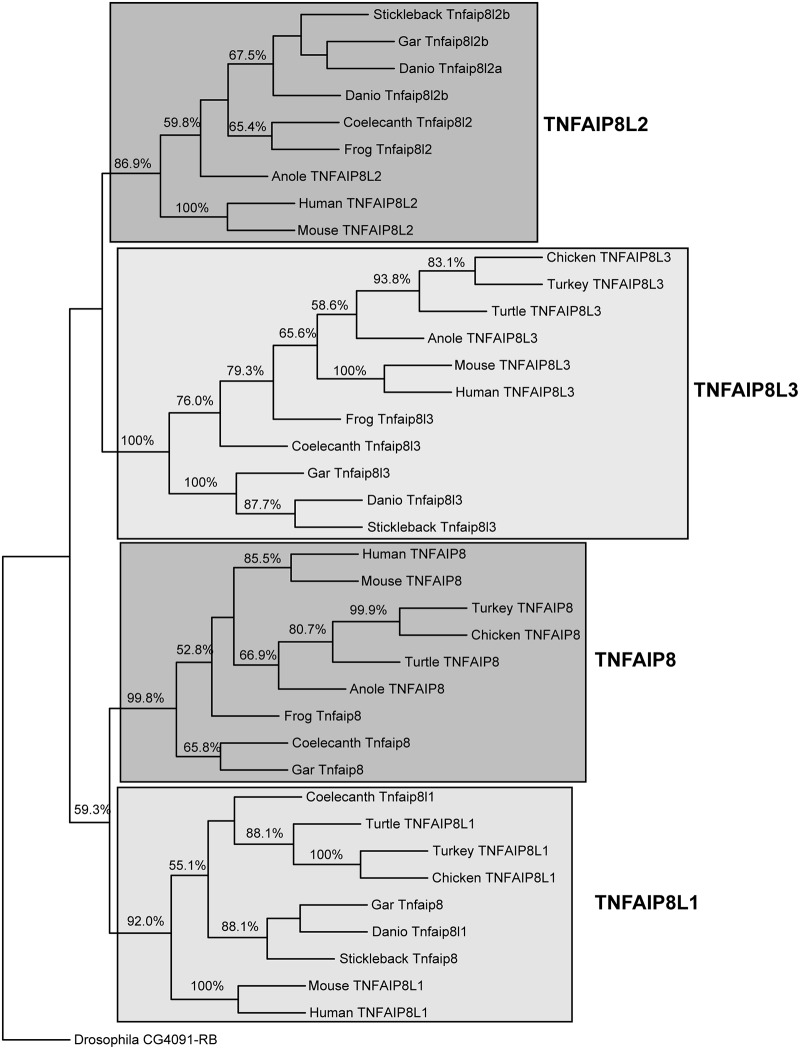
Fig 8. Phylogeny of the TNFAIP8 family of proteins.
Sequences were collected from Ensembl (www.ensembl.org) and GenPept (http://www.ncbi.nlm.nih.gov/protein) and aligned using Clustal Omega (S2 Fig) [30]. Sequence identifiers are provided in S2 Fig. Drosophila sequence CG4091-RB was used as an outgroup. The unrooted consensus tree with % bootstrap support for each node (above 50%) is presented. The common and binomial names of animals comprising the alignment (S2 Fig) and cladogram are as follows: zebrafish (Danio rerio), mouse (Mus musculus), human (Homo sapiens), frog (Xenopus tropicalis), chicken (Gallus gallus) and turkey (Meleagris gallopavo), stickleback (Gasterosteus aculeatus), spotted gar (Lepisosteus oculatus), coelacanth (Latimeria chalumnae), Chinese softshell turtle (Pelodiscus sinensis) and anole lizard (Anolis carolinensis).