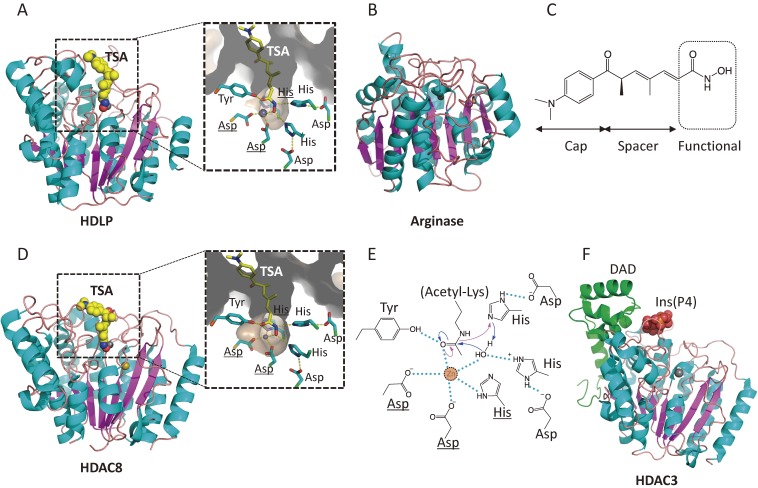
Figure 4.
Crystal structures and catalytic mechanisms of HDAC proteins. (A) HDLP (PDB id: 1C3R). TSA and zinc ion are represented as space-filled spheres, in which C, N, O, and Zn atoms are colored in yellow, blue, red, and grey, respectively. Close-up view of the active site is shown in the right panel. Two asparagine residues and histidine (underlined) coordinate the active site Zn2+ ion. (B) Arginase (1RLA). (C) The chemical structure of TSA, consisting of a cap, a spacer, and a zinc-binding functional group. (D) HDAC8 (1T64). K+ ion (Na+ ion in this structure due to the crystallization conditions) is colored in khaki. (E) Catalytic mechanism of class I HDACs. The catalytic histidine residue facilitates the nucleophilic attack at the substrate carbonyl by activating a water molecule (blue arrow). Hydrogen bond interactions are drawn in dotted lines. (F) HDAC3 in complex with inositol-tetraphosphate and DAD (4A69). The inositol phosphate is represented as space-filled spheres, in which C, O, and P atoms are depicted in yellow, red, and orange, respectively. The DAD is colored in green.