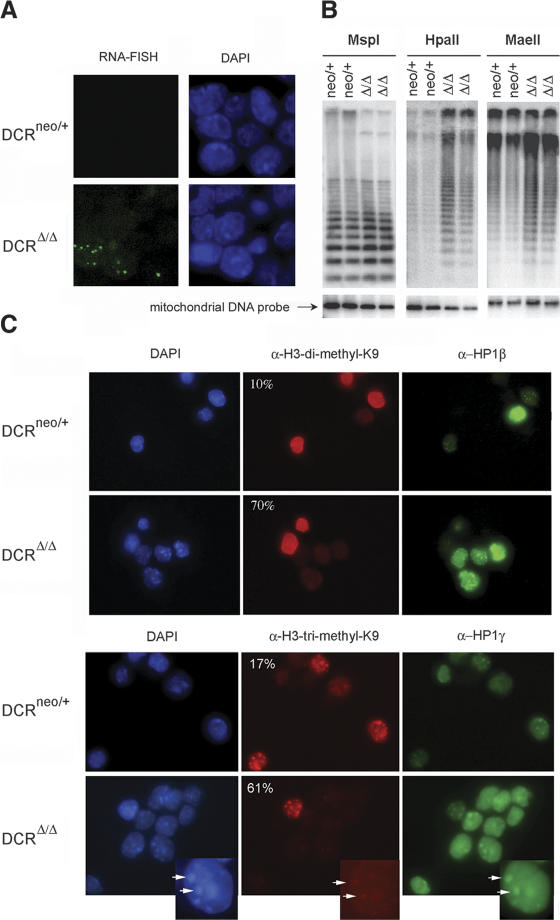
Figure 4.
Evidence for transcriptional derepression of centromeric repeats. (A) RNA FISH using a centromeric DNA probe was performed on DCRneo/+ and DCRΔ/Δ ES cells. The respective DAPI nuclear staining is shown next to each panel (63×). (B) DNA methylation analysis of genomic DNA derived from two DCRneo/+ and two DCRΔ/Δ clones. Equivalent amounts of DNA were digested with MspI, HpaII, or MaeII. (Lower panel) EtBr staining of the gel (data not shown) and rehybridization with a mitochondrial DNA probe demonstrate equivalent loading. MspI cleaves irrespective of whether the CpG dinucleotide within the cognate restriction site is methylated, whereas HpaII or MaeII will not. Gels were blotted on a nitrocellulose membrane and hybridized with a probe specific for the minor satellite repeat. (C) Histone modifications and HP1 protein localization are altered in Dicer-deficient cells. DCRneo/+ and DCRΔ/Δ cells were stained either with a rabbit polyclonal antibody specific for dimethyl-H3K9 and a monoclonal antibody against HP1β (upper panels) or a polyclonal antibody specific for trimethyl-H3K9 and a monoclonal antibody against HP1γ (lower panels). DAPI staining of nuclei for each panel is shown on the left (63× magnification). Percentages of cells unstained or dim for variously methylated H3K9 species are indicated on the top left corner of each panel. At least 200 cells from each clone were counted. Inset figures point to an example of the colocalization of HP-1γ, trimethyl H3K9 staining, and centromeric heterochromatin as reflected by the existence of DAPI dense spots.