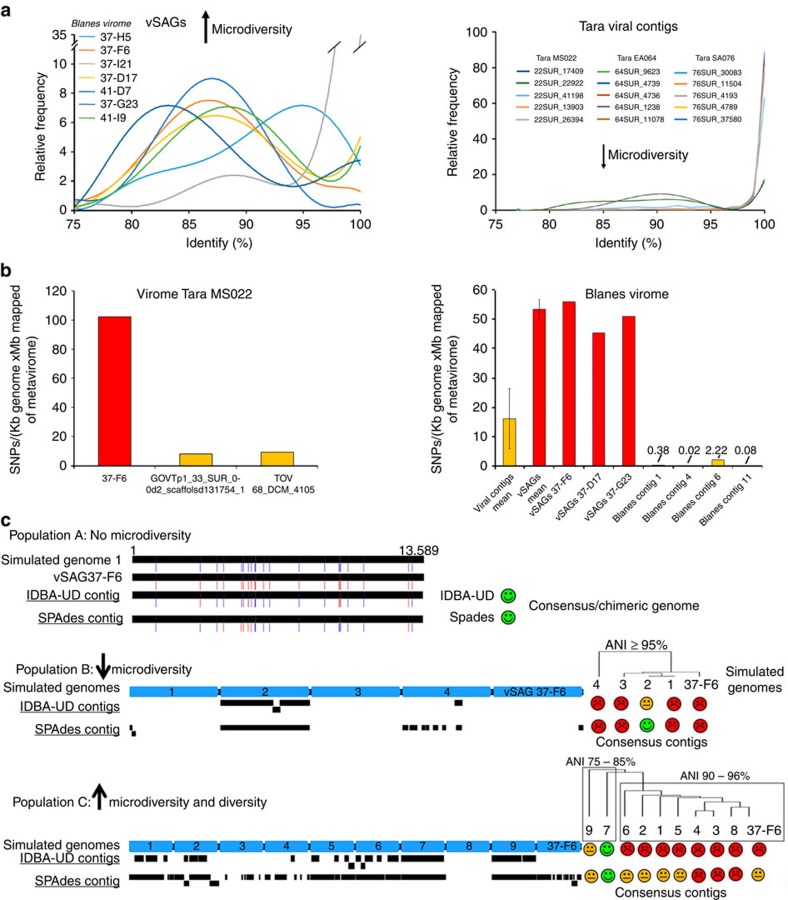
Figure 6. Assessment of natural vSAGs microdiversity and impact on metagenomic assembly.
(a) Species-specific recruitment patterns (also referred as diversity curves) for vSAGs and highly abundant viral contigs from viromics. Curves represent the percentage of recruited reads (Y axis) at different nucleotide identity values (X axis) for vSAGs and Tara Oceans contigs3 in their own viromes. The five most recruiting viruses of each viral data set are shown for convenience. (b) SNP frequency for most abundant viral populations at the species level (≥95% nucleotide identity) of vSAGs and viral contigs (within the top 30 ranking in recruitment) recovered by viromics from the Blanes Bay Microbial Observatory (same sampling site of surface vSAGs) and the Tara Mediterranean MS022 data set3. In Blanes Bay Microbial Observatory, mean±s.d. of most abundant viral contigs (25 contigs) and vSAGs (4 contigs) are shown. (c) Impact of viral diversity and microdiversity on genome reconstruction by metagenomics. Three populations of virus 37-F6 with different (micro)-diversities were simulated within the virome Tara MS022 (ref. 3) (see details in Supplementary Fig. 20 and Supplementary Note 5). Population A lacked microdiversity (two simulated nearly identical genomes of 37-F6 with 20 SNPs). A chimeric contig with a mixture of SNPs was obtained (SNPs in blue from simulated genome 1, and in red from vSAG 37-F6). Population B simulated a simplistic scenario with five genomes (ANI≥95%) without high genetic variability in the hypervariable genomic island (Fig. 4; Supplementary Fig. 14). SPAdes assembler reconstructed a consensus contig from only one of the simulated genomes. Population C simulated a more realistic microdiverse scenario than observed in panel A with 10 simulated co-existing viruses (ANI 75-95% and high variability in the genomic island (see details in Supplementary Fig. 20 and Supplementary Note 5). The genome was almost entirely assembled only from those distantly related viruses 7 and 9, while 37-F6 genome could not be assembled. Blue arrows depict the simulated genomes. Black blocks depict the resulting assembled contigs by IDBA_UD and SPAdes assemblers.