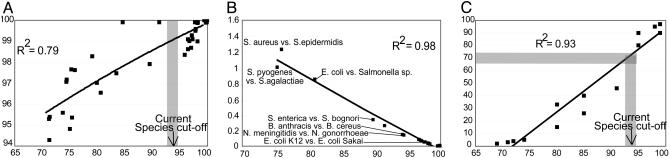
Fig. 1.
Relationships between ANI, 16S rRNA, mutation rate, and DNA–DNA reassociation. Each black square represents the ANI of all conserved genes between two strains (x axes) plotted against (y axes) the 16S rRNA sequence identity (A), the average rate of synonymous nucleotide substitutions (B), and the DNA–DNA reassociation values (C) of the two strains. The shaded bar represents 93–94% ANI, which approximately corresponds to 70% DNA–DNA reassociation value, i.e., the species cutoff for prokaryotic species, according to the regression analysis in C. Shown in B is the average rate of synonymous substitutions for all genes in the genome. Comparable results were also obtained when analysis was restricted to genes with no apparent codon biases, or to only fourfold-degenerated sites, as opposed to all sites in a gene (data not shown).