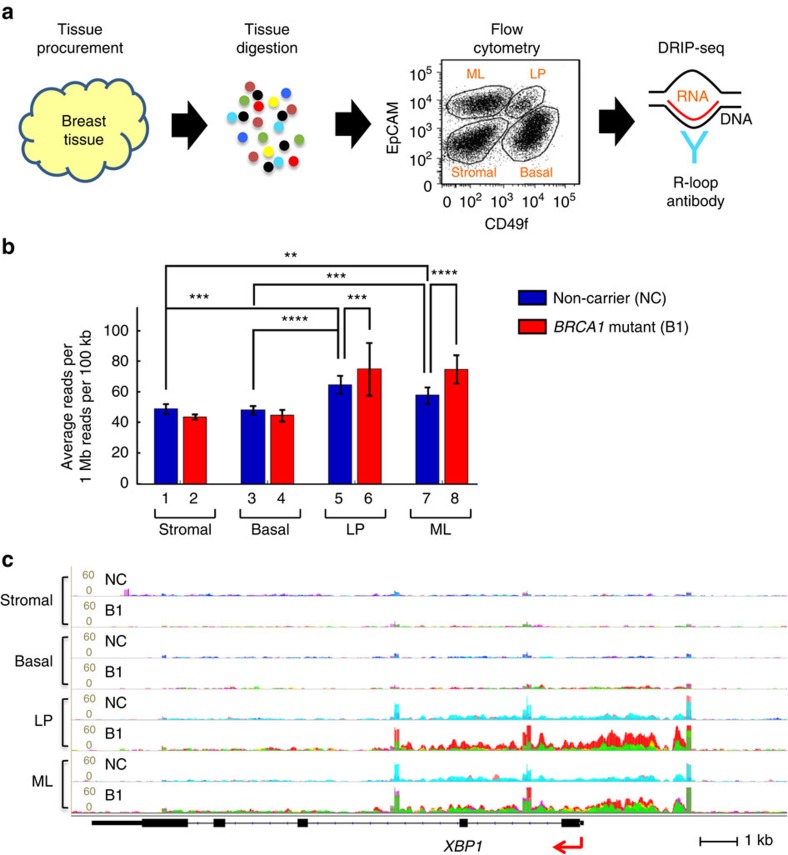
Figure 2. DRIP-seq validation of luminal lineage-specific R-loop accumulation in BRCA1 mutation carriers.
(a) Experimental design for DRIP-seq. Fresh cancer-free breast tissue samples were digested into single cells, and then sorted by flow cytometry into four populations: stromal, basal, LP and ML. Genomic DNA from each population was extracted, digested and immunoprecipitated with an R-loop-specific antibody S9.6. DRIP DNA was subjected to deep sequencing. (b) Average reads per 1 Mb reads per 100 kb (from TSS to TTS) in stromal–basal–LP–ML population in non-carriers (n=4) and BRCA1 mutation carriers (n=4). **P≤0.01, ***P≤0.001 and ****P<0.0001 by permutation test. Error bars represent s.e.m. (c) Track view of DRIP-seq density profile centred on gene XBP1. Each track is an overlay of four individual non-carriers (NC) or four BRCA1 mutation carriers (B1) indicated by different colours. TSS was marked by red arrow.