Fig. 8.
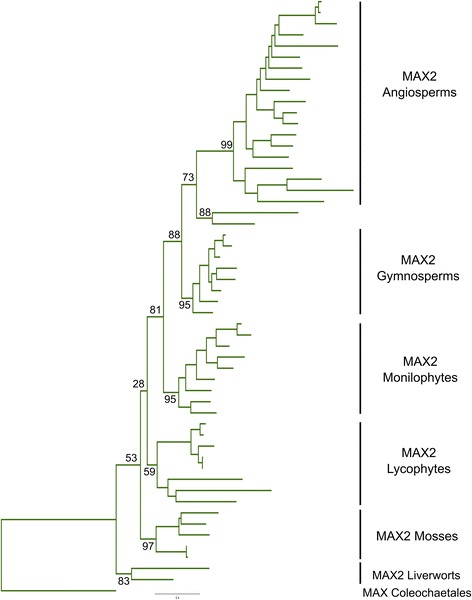
The MAX2 family has a very conservative evolutionary history. Codon-level phylogenetic analysis implemented in GARLI on the whole MAX2 family (57 sequences from 54 species). This analysis was performed using an optimized character set (see Methods). Phylogram showing the ‘most likely’ tree from GARLI analysis, labelled to show the high-order relationships between the major clades (as described in Table 1).Trees were rooted with charophyte sequences, consistent with contemporary notions of plant organismal phylogeny. Numbers associated with internal branches denote maximum likelihood bootstrap support (percent support)
