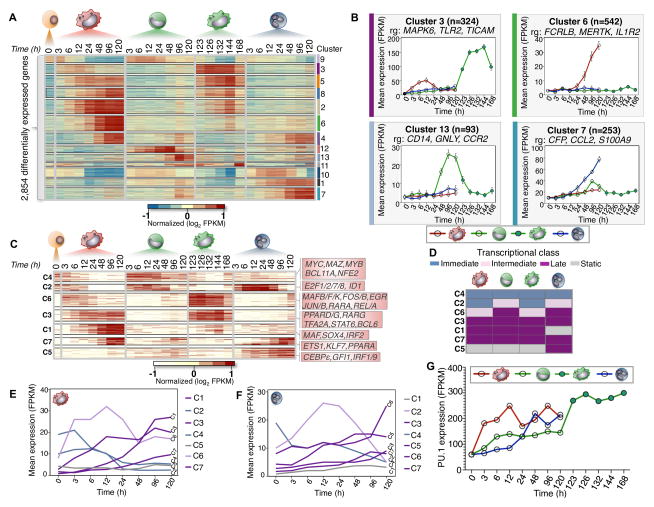
Figure 3. Temporal modules of transcriptional regulator expression.
(A) Heatmap of 2,854 differentially expressed genes during myeloid differentiation (α=0.05, FDR < 1%, p-val < 0.01). 13 expression clusters were derived using k-means and denoted by both color and number for all differentially expressed genes. FPKM values log transformed and row-mean normalized for all genes. (B) Gene expression profiles for clusters (3, 6, 7, and 13). Mean expression and standard deviation (error bars) are plotted across all biological replicates and for all genes in each corresponding cluster. Cluster size (n) and representative genes (rg) are denoted. (C) Heatmap of 232 differentially expressed transcriptional regulators (α=0.05, FDR < 1%, p-val < 0.01). Seven regulator clusters were derived using k-means (C1–C7). Representative regulators from each cluster are shown respectively. FPKM values are log transformed and row-mean normalized for all regulators. (D) Schematic of transcriptional regulator classification. Transcriptional classes are denoted as immediate (blue), intermediate (pink), late (Purple) and static (Gray). (E and F) Transcriptional profiles for each cluster based on classification for macrophage and neutrophil cells, respectively. Mean FPKM expression is shown for each cluster. (G) PU.1 gene expression during myeloid differentiation increases across all cell-types. The mean FPKM values for each time-series are shown.