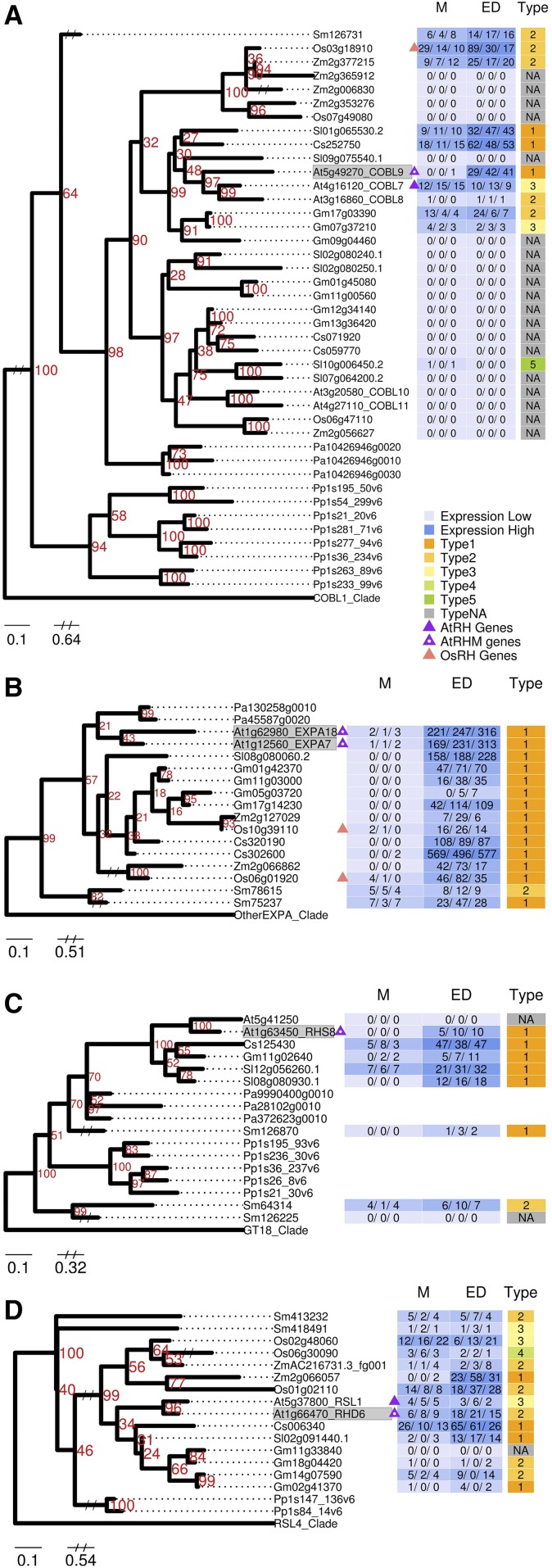
Figure 6.
Representative maximum likelihood phylogenetic trees of AtRHM gene families. A, COBL9. B, EXPA7. C, RHS8. D, RHD6. For each tree, the defining Arabidopsis AtRHM gene(s) associated with a mutant phenotype is shaded in gray. Gene expression FPKM values are shown for each replicate and converted to a heat map with high expression in darker blue and low expression in light blue. Expression profile types generated from the fold change between two developmental zones are shown in different colors as indicated in the key. Triangles indicate AtRHM genes (purple with white dot), AtRH genes (solid purple), and OsRH genes (pink). Numbers in red indicate support levels from 1,000 bootstrap. Gene identifiers are abbreviated (Arabidopsis, At; cucumber, Cs; soybean, Gm; rice, Os; tomato, Sl; Selaginella, Sm; maize, Zm; P. patens, Pp; Norway spruce, Pa). M, Meristematic zone; ED, combined elongation plus differentiation zones; Type, expression profile types. The complete set of 14 AtRHM family trees is presented in Supplemental Figure S3.