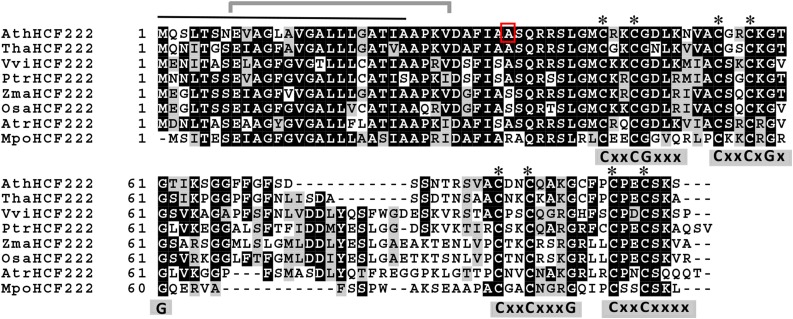
Figure 5.
Alignment of HCF222 orthologs from seed plants and a bryophyte. Multiple amino acid sequence alignment of Arabidopsis HCF222 (AtHCF222) and homologs from Tarenaya hassleriana (Tha), Vitis vinifera (Vvi), Zea mays (Zma), Oryza sativa (Osa), Populus trichocarpa (Ptr), Amborella trichopoda (Atr), and the bryophyte M. polymorpha (Mpo). Identical amino acids are marked white on black; similar amino acids are black on gray. The predicted signal peptide of HCF222 is marked by a black line on top, the gray bracket indicates the predicted membrane-spanning helix, and the red frame indicates the mutated amino acid (A→V) in the mutant allele hcf222-1. Asterisks mark conserved Cys residues, and, in addition, the four CxxC motifs and conserved Gly residues are shown below the sequence. The amino acid sequence alignment was generated using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) and Boxshade 3.21 (http://www.ch.embnet.org/software/BOX_form.html).