Magnesium transported by OsMGT1 in rice roots is required for enhancing transport activity of OsHKT1;5, which confers salt tolerance through retrieval of Na from xylem vessels.
Abstract
Salt stress is one of the major factors limiting rice (Oryza sativa) production globally. Although several transporters involved in salt tolerance have been identified in rice, the mechanisms regulating their transport activity are still poorly understood. Here, we show evidence that a rice Mg transporter OsMGT1 is required for salt tolerance probably by regulating transport activity of OsHKT1;5, a key transporter for the removal of Na+ from the xylem sap at the root mature zone. Knockout of OsMGT1 did not affect total Na uptake, but increased Na concentration in the shoots and xylem sap, resulting in a significant increase in salt sensitivity at low external Mg2+ concentration (20–200 μm). However, such differences were abolished at a higher Mg2+ concentration (2 mm), although the total Na uptake was not altered. OsMGT1 was expressed in both the roots and shoots, but only that in the roots was moderately up-regulated by salt stress. Spatial expression analysis revealed that OsMGT1 was expressed in all root cells of the root tips but was highly expressed in the pericycle of root mature zone. OsMGT1 was also expressed in the phloem region of basal node, leaf blade, and sheath. When expressed in Xenopus laevis oocytes, the transport activity of OsHKT1;5 was enhanced by elevating external Mg2+ concentration. Furthermore, knockout of OsHKT1;5 in osmgt1 mutant background did not further increase its salt sensitivity. Taken together, our results suggest that Mg2+ transported by OsMGT1 in the root mature zone is required for enhancing OsHKT1;5 activity, thereby restricting Na accumulation to the shoots.
Soil salinity is a major abiotic stress limiting the yield of agricultural crops worldwide. Approximately 7% of the world’s total land area is affected by high salt stress (Shabala and Cuin, 2008; Munns and Tester, 2008). High salt concentration inhibits plant growth through osmotic stress and ionic Na+ stress (Munns and Tester, 2008; Horie et al., 2012). Osmotic stress causes inhibitions of water uptake, cell elongation, and leaf development, while ionic stress results in high Na accumulation in the shoots, which decreases protein synthesis, enzymatic reactions, and photosynthetic processes (Zhu, 2001; Horie et al., 2012; Deinlein et al., 2014). Therefore, restricting Na accumulation to the shoots (especially in the leaves) is important to protect plants from ionic Na+ stress (Davenport et al., 2005; Yamaguchi et al., 2013).
Rice (Oryza sativa) is the most salt-sensitive species among cereals (Munns and Tester, 2008; Lutts et al., 1996), but several genes involved in salt tolerance in rice have been identified. OsSOS1 in rice was isolated based on the homology with AtSOS1 in Arabidopsis (Arabidopsis thaliana; Martínez-Atienza et al., 2007; Shi et al., 2000). Similar to AtSOS1, OsSOS1 also encodes a plasma membrane-localized Na+/H+ antiporter, which mediates active Na+ extrusion in the roots under salt stress (Shi et al., 2000). Although the phenotype of OsSOS1 knockout lines under salt stress is unknown, OsSOS1 was able to complement salt tolerance of atsos1 mutant, indicating that OsSOS1 also plays an important role in salt tolerance in rice (Martínez-Atienza et al., 2007). Furthermore, the CBL-interacting protein kinases OsCIPK24 and the calcineurin B-like protein OsCBL4, which are homologs of AtSOS2 and AtSOS3 in Arabidopsis, respectively, act coordinately to activate OsSOS1 transport activity. This SOS-mediated pathway may represent a basic salt tolerance in both dicots and monocots. After Na+ uptake, a fraction of Na+ taken up is sequestered into the vacuoles of both root and shoot cells by AtNHX1, a vacuolar Na+, K+/H+ exchanger in Arabidopsis (Apse et al., 1999, 2003; Leidi et al., 2010). Its homolog in rice has also been implicated in salt tolerance, although the exact role is not well understood (Chen et al., 2007).
Several members of HKT gene subfamily in rice, including OsHKT1;1, OsHKT1;3, OsHKT1;4, and OsHKT1;5 have also been implicated in salt tolerance in rice (Hamamoto et al., 2015). All these members function as a Na+ influx transporter but have different expression patterns. OsHKT1;1 encodes a plasma membrane-localized protein and was mainly expressed in the phloem of leaf blades (Wang et al., 2015). Knockout of this gene resulted in increased salt sensitivity and Na accumulation in the shoots, indicating that OsHKT1;1 is involved in retrieving Na+ from the leaf blade (Wang et al., 2015). Furthermore, its expression was regulated by an MYB-type transcription factor (OsMYBc). By contrast, OsHKT1;3 was localized to the Golgi (Rosas-Santiago et al., 2015). It is expressed in the vascular tissues of roots and leaves (Jabnoune et al., 2009), but its exact role in salt tolerance is unknown. OsHKT1;4 is mainly expressed in the leaf sheath and encodes a plasma membrane-localized protein (Suzuki et al., 2016). Recent functional analysis showed that OsHKT1;4 does not contribute to salt tolerance at the vegetative growth stage (Suzuki et al., 2016); however, at the reproductive stage, knockdown of OsHKT1;4 resulted in increased Na accumulation in the leaf sheath and leaf blade under salt stress, implicating that this gene may be involved in the Na+ exclusion in the leaf sheath (Suzuki et al., 2016). On the other hand, SKC1/OsHKT1;5 was suggested to be a quantitative trait locus controlling a higher K+/Na+ ratio in the shoots (Ren et al., 2005). In contrast to OsHKT1;4, SKC1/OsHKT1;5 is highly expressed in the roots (Ren et al., 2005). Furthermore, it is preferentially expressed in the parenchyma cells surrounding the xylem vessels. Similar to AtHKT1;1 in Arabidopsis (Mäser et al., 2002; Sunarpi et al., 2005) and TmHKT1;5-A and TaHKT1;5-D in wheat (Triticum aestivum; James et al., 2006, 2011; Byrt et al., 2014), SKC1/OsHKT1;5 is therefore thought to be responsible for retrieving Na+ from the xylem sap, leading to low Na accumulation in the shoots (Ren et al., 2005; Deinlein et al., 2014). All these studies show that these HKT genes play important roles in salt tolerance in different organs and tissues of rice; however, the mechanisms regulating the transport activity of HKT proteins are still poorly understood.
MGT family proteins have been known as Mg transporters in both prokaryote and eukaryote (Hmiel et al., 1986; Li et al., 2001). There are 10 MGT homologs in the Arabidopsis genome and nine homologs in rice (Schock et al., 2000; Gebert et al., 2009). Among them, AtMGT6 in Arabidopsis and OsMGT1 in rice mediate root Mg uptake, respectively, although they differ in their gene expression patterns (Mao et al., 2014; Chen et al., 2012). In this study, we found that OsMGT1 is involved in enhancing OsHKT1;5 transport activity at least in rice. OsMGT1 is a plasma membrane-localized transporter for Mg2+ (Chen et al., 2012; Chen and Ma, 2013). It was initially found to be involved in Al tolerance. Rapid up-regulation of OsMGT1 expression in response to Al stress resulted in increased cytosolic Mg concentration, thereby preventing Al binding to enzymes and other cellular components (Chen et al., 2012; Chen and Ma, 2013). Further detailed analysis of this gene revealed that OsMGT1 is also required for conferring salt tolerance in rice through enhancing the transport activity of OsHKT1;5, a key player of salt tolerance in rice.
RESULTS
Knockout of OsMGT1 Resulted in Hypersensitivity to Salt Stress
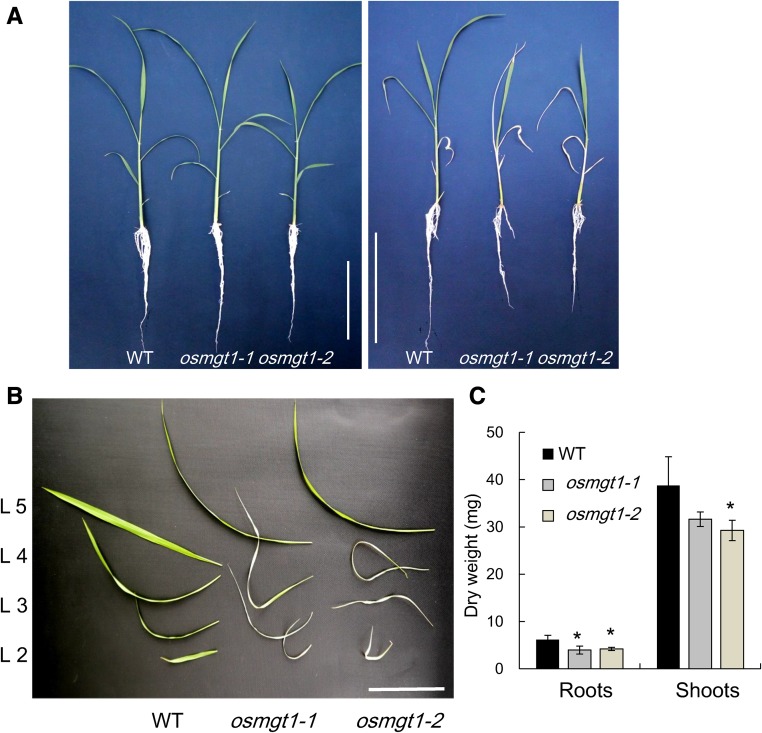
To investigate the role of OsMGT1 in salt tolerance in rice, we grew both the wild-type rice and two OsMGT1 knockout lines in a nutrient solution containing 0.27 mm Mg2+ in the presence or absence of 50 mm NaCl. In the absence of NaCl, the growth was similar between the two independent OsMGT1 knockout lines (osmgt1) and wild type (Fig. 1A). However, in the presence of 50 mm NaCl, the old leaves of osmgt1 mutants were wilt after exposure to 12 d (Fig. 1, A and B), but that of wild type was hardly affected. The dry weight of the roots and shoots was 34% and 22% lower in the mutants than in the wild type (Fig. 1C).
Figure 1.
Sensitivity of OsMGT1 knockout lines to salt stress. A, Effect of salt stress on the growth of rice seedlings. Left, 0 mm NaCl; right, 50 mm NaCl. B, Phenotype of each leaf under salt stress. C, Dry weight of roots and shoots. Seedlings of both wild-type rice (WT) and two OsMGT1 knockout lines (osmgt1-1/NF0595 and osmgt1-2/NE45208) were exposed to a nutrient solution containing 50 mm NaCl. After 12 d, individual leaves were separated and photographed. Data are means ± sd (n = 3). Bar = 10 cm. The asterisk shows a significant difference compared with wild type (P < 0.05 by Tukey’s test).
To examine the possibility that the decreased growth in the mutants is caused by osmotic stress, we grew them in a nutrient solution containing high concentrations of mannitol. Although the growth was inhibited by high concentration of mannitol, there was no difference in the growth between the wild type and mutants (Supplemental Fig. S1A). There was also no difference in the water loss rate of detached leaves (Supplemental Fig. S1B). These results indicate that decreased salt tolerance in osmgt1 mutants is caused by Na+ toxicity rather than osmotic stress. In addition, we also investigated the sensitivity of osmgt1 mutants to high Mg2+ (10 mm) and Cd2+ (5 µm) toxicity (Supplemental Fig. S2). High Mg and Cd inhibited the growth of both wild type and osmgt1 mutants, but there was no difference in the tolerance to them between wild type and mutants, indicating that OsMGT1 plays a specific function in Na+ tolerance.
osmgt1 Mutant Showed Higher Na Accumulation in the Shoots
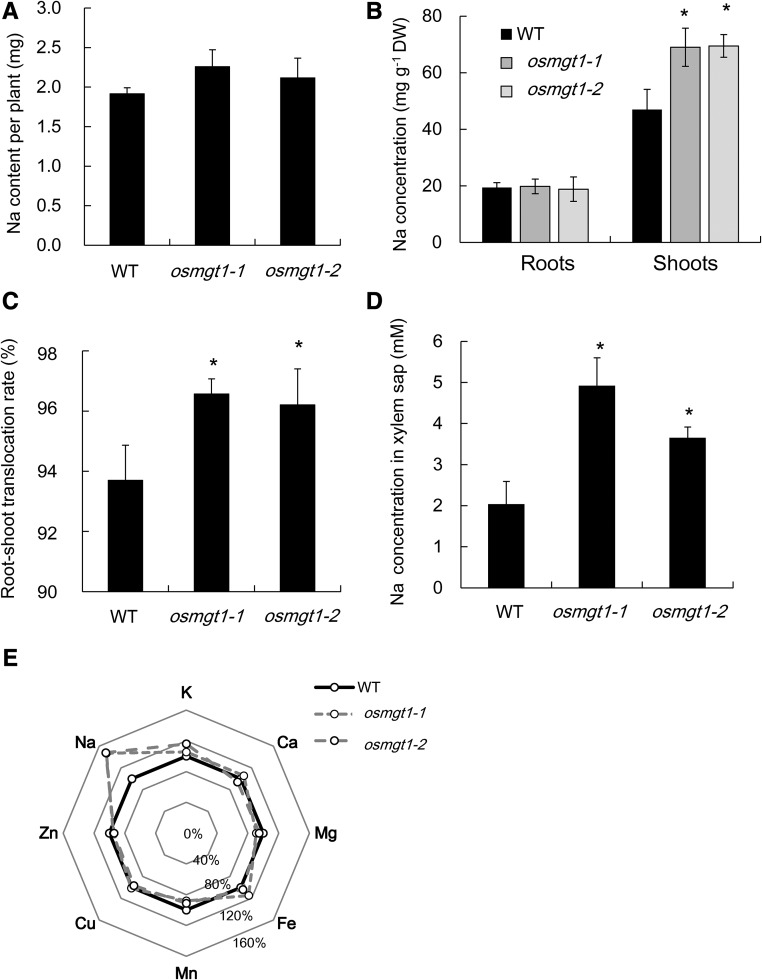
The total Na uptake (the total Na content per plant) was similar between the mutants and wild type (Fig. 2A). However, Na concentration of the shoots was 1.5-fold higher in the mutants than in the wild type, although that of the roots was similar (Fig. 2B). As a result, the root-to-shoot translocation of Na (the percentage of shoot Na content in total Na content in plant) was 96% in the mutants, in contrast to 93% in the wild type (Fig. 2C). The Na concentration in xylem sap was also higher in the mutants than in the wild type (Fig. 2D). By contrast, Mg concentration in mutant roots was lower than wild type, although that in the shoots was similar (Supplemental Fig. S3A). K concentration in the roots and shoots was similar between the mutant and wild type (Supplemental Fig. S3B). There was also no difference in Mg and K concentration in the xylem sap between wild type and mutants (Supplemental Fig. S3, C and D). Furthermore, no difference was found in the concentration of other mineral elements in both the roots and shoots between wild type and mutants (Fig. 2E).
Figure 2.
Comparison of Na and other mineral elements in the shoots and roots of wild-type rice and knockout lines of OsMGT1. A, Total Na content per plant. B, Na concentration of the roots and shoots. C, Root-to-shoot translocation rate of Na. Seedlings of both wild-type (WT) and two OsMGT1 knockout lines (osmgt1-1 and osmgt1-2) were exposed to a nutrient solution containing 50 mm NaCl for 12 d. D, Na concentration in xylem sap. Xylem sap was collected from the wild type and the knockout lines exposed to a nutrient solution containing 10 mm NaCl for 24 h. E, Percentage difference of Na and other seven mineral elements in the shoots of osmgt1 mutants compared with the wild type. Data are means ± sd (n = 3). The asterisk shows a significant difference compared with wild type (P < 0.05 by Tukey’s test).
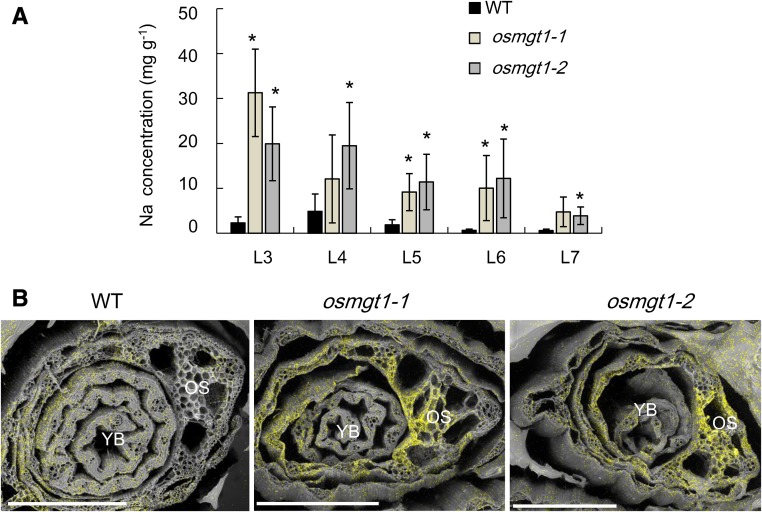
Leaf age-dependent analysis showed that Na accumulation in the leaf blade increased with the age in both the wild type and mutants (Fig. 3A), but the mutants always accumulated higher Na than the wild type in each leaf. Furthermore, with the help of the scanning electron microscopy (sem) with energy dispersive x-ray spectroscopy (EDS), a stronger signal of Na was detected in the old leaves of the mutants than those of the wild type (Fig. 3B).
Figure 3.
Leaf age-dependent Na accumulation in the wild-type rice and knockout lines of OsMGT1. A, Na concentration in different leaves. Rice seedlings were exposed to a nutrient solution containing 50 mm NaCl for 3 d. The leaf blades 3 to 7 (from old to young) were separately sampled. Data are means ± sd (n = 3). The asterisk shows a significant difference compared with wild type (WT; P < 0.05 by Tukey’s test). B, Na accumulation and distribution by SEM and EDS. Seedlings were exposed to 50 mm NaCl for 7 d, and the shoot section at 10 mm from the basal region was excised. The scanning electronic photos and elemental distribution photos were generated by SEM and EDS, respectively. YB, young leaf blade; OS, old leaf sheath. Yellow color indicates Na. A representative image of six shoot samples is presented. Bar = 500 μm.
High Mg2+ Restored the Salt Tolerance of osmgt1 Mutants
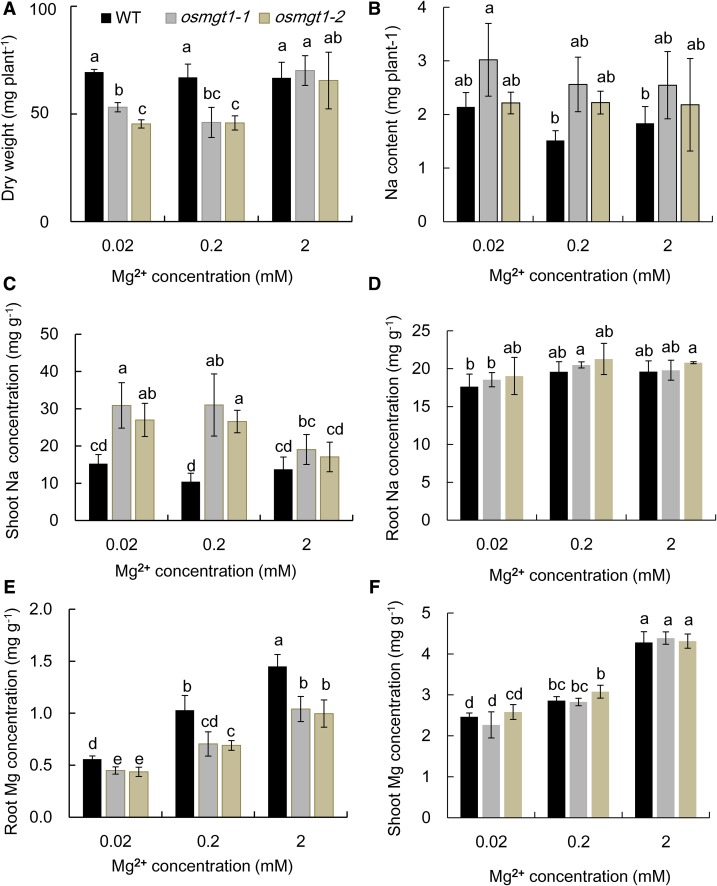
To investigate whether salt toxicity in the knockout lines can be alleviated by Mg2+, we exposed the seedlings to a solution containing low (0.02, 0.2 mm) and high (2 mm) Mg2+ in the presence of 50 mm Na+. At low external Mg2+ concentrations, the growth of osmgt1 mutants was significantly inhibited compared with wild type (Fig. 4A). However, at high Mg2+ condition, the growth of osmgt1 mutant was restored to the level similar to the wild type (Fig. 4A). The total Na uptake was not altered by increasing external Mg2+ concentrations (Fig. 4B), and there was also no difference between the wild type and mutants. The Na concentration in the shoots of osmgt1 mutants was higher than that in the wild type at low Mg2+ concentrations but was decreased by nearly 50% at high Mg2+ concentration (Fig. 4C) and became similar to that of wild type at 2 mm Mg. By contrast, the Na concentration in the shoots of wild type was not affected by external Mg2+ concentrations (Fig. 4C). The Na concentration in the roots was similar between wild type and mutants at all Mg2+ concentrations and was not affected by Mg2+ concentrations (Fig. 4D). We also compared Mg concentration of wild type and osmgt1 mutants. The Mg concentration in the roots increased with increasing Mg2+ supply in all lines, although osmgt1 mutants always showed a significantly lower Mg concentration than wild type at each Mg2+ supply (Fig. 4E). However, at 2 mm Mg, Mg concentration in the roots of osmgt1 mutants reached to a similar level of the wild type at 0. 2 mm Mg (Fig. 4E). By contrast, there was no difference in the shoot Mg concentration between wild type and mutants, although it was also increased with increasing external Mg2+ concentrations (Fig. 4F).
Figure 4.
Alleviation of salt stress by high Mg2+ supply in OsMGT1 knockout lines. A and B, Dry weight (A) and total Na content (B) per plant. C and D, Na concentration in shoots (C) and roots (D). E and F, Mg concentration in roots (E) and shoots (F). Seedlings of both wild-type rice (WT) and two OsMGT1 knockout lines (osmgt1-1 and osmgt1-2) were exposed to a nutrient solution containing 0.02, 0.2, or 2 mm MgCl2 in the presence of 50 mm NaCl for 7 d. Data are means ± sd (n = 3). Means with different letters are significantly different (P < 0.05 by Tukey’s test).
OsMGT1 Expression Was Moderately Up-Regulated in Response to Salt Stress
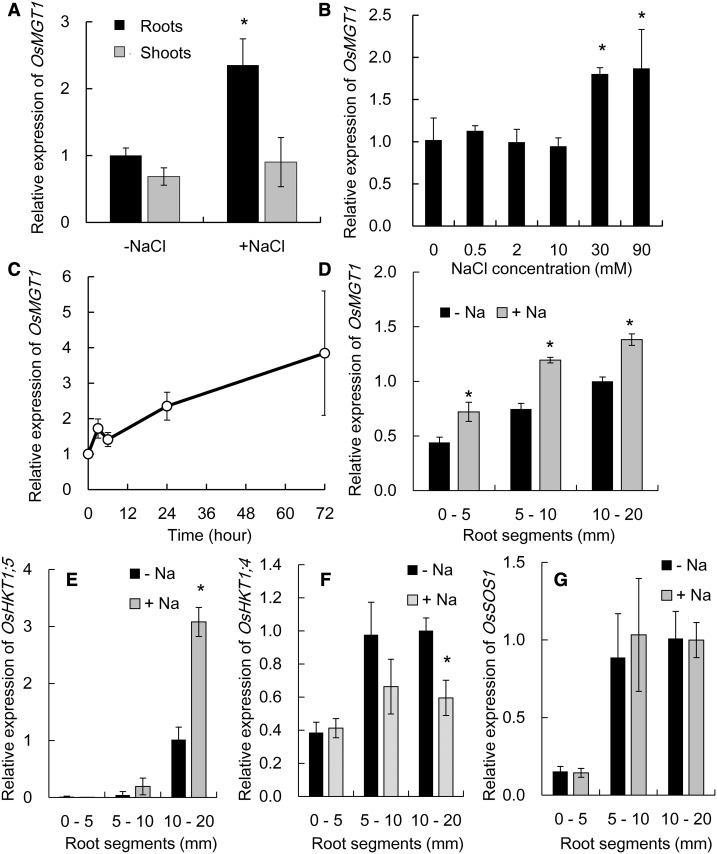
OsMGT1 was expressed in both the shoots and roots, but only that in the roots was up-regulated by about 2-fold after exposure to 50 mm NaCl for 24 h (Fig. 5A). A dose-response experiment showed that OsMGT1 expression was not induced by low external NaCl concentrations (<10 mm) but induced by high NaCl concentrations (>30 mm; Fig. 5B). A time-course experiment showed that the induction of OsMGT1 occurred at 3 h after the exposure to 50 mm NaCl and the expression was increased with exposure times (Fig. 5C).
Figure 5.
Gene expression pattern of OsMGT1 in response to salt stress. A, Expression of OsMGT1 in the roots and shoots. Rice seedlings were exposed to a solution containing 50 mm NaCl for 24 h. B, Dose-response expression of OsMGT1 in rice roots. Rice seedlings were exposed to a solution containing different NaCl concentrations for 6 h. C, Time-dependent expression of OsMGT1 in rice roots. Rice seedlings were exposed to a solution containing 50 mm NaCl for different time. D to G, Root spatial expression pattern of OsMGT1 (D), OsHKT1;5 (E), OsHKT1;4 (F), and OsSOS1 (G). Different segments (0–5, 5–10, and 10–20 mm from root tips) were excised after the roots exposed to 0 or 50 mm NaCl for 24 h. The expression level was determined by real-time RT-PCR. Data are means ± sd (n = 3). The asterisk shows a significant difference compared with −Na in A, B, and D to F (P < 0.05 by Tukey’s test).
Spatial expression analysis revealed that the expression of OsMGT1 was higher in the root mature zones than in the root tips, but the expression in all root regions was up-regulated by NaCl (Fig. 5D). We also compared root spatial expression pattern of some salt tolerance genes. OsHKT1;5 was highly expressed in the root mature zone and its expression was up-regulated by NaCl exposure (Fig. 5E). By contrast, OsHKT1;4 was down-regulated by NaCl (Fig. 5F), and the expression of OsSOS1 was not affected by NaCl exposure (Fig. 5G). The expression of both OsHKT1;4 and OsSOS1 was also higher in the mature root region than the root tips (Fig. 5, F and G). In the osmgt1 mutant, the expression of OsHKT1;4, OsHKT1;5, and OsSOS1 was not altered compared to the wild type under both salt and nonsalt stress (Supplemental Fig. S4).
Tissue Specificity of Expression of OsMGT1
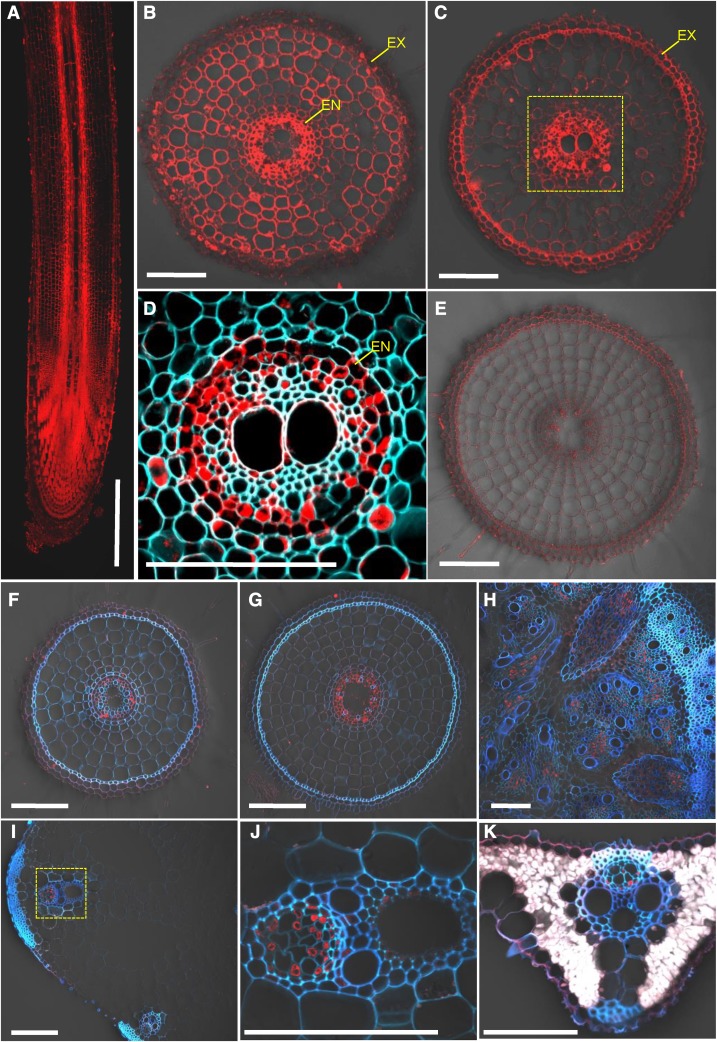
To examine the tissue and cell specificity of OsMGT1 expression, we generated transgenic rice lines carrying the 2.5 kb promoter sequence of OsMGT1 fused with GFP. Immunostaining of the transgenic rice with a GFP antibody showed that the signal was detected in the whole root, but with different patterns (Fig. 6). In the root tips, the signal was observed in all root cells (Fig. 6, A and B), whereas in the root mature zone, strong signal was only detected in the stellar tissues, especially in the pericycle (Fig. 6, C and D). No signal was detected in the roots of wild-type rice (Fig. 6E), indicating high specificity of this antibody. The signal in the root mature zone was enhanced by salt stress (Fig. 6, F and G). In addition, the signal was also observed in the phloem region of basal node (Fig. 6H), leaf sheath (Fig. 6, I and J), and leaf blade (Fig. 6K).
Figure 6.
Tissue-specific and Na-responsive expression of OsMGT1. Ato E, Expression of OsMGT1 in different root region. Immunostaining with an anti-GFP antibody was performed in different root zone of pOsMGT1-GFP transgenic rice (A–D) and wild-type rice (E), including longitudinal section of root (A), cross sections at 5 mm (B and E), and 30 mm (C and D) from root apex. F to K, Expression of OsMGT1 expression to salt stress. Immunostaining of root segments (20 mm from root tips; F and G), basal node (H), leaf sheath (I and J), and leaf blade (K), after exposure to 0 (F) or 50 mm (G–K) NaCl for 24 h. Red color shows signal from GFP antibody detected with a secondary antibody. Cyan color shows cell wall autofluorescence. Yellow-dotted areas in C and I were magnified in D and J, respectively. EN, Endodermis; EX, exodermis. Five independent transgenic lines were investigated, and the representative results are shown. Bars = 500 μm (A) and 100 μm (B–K).
Mg2+ Enhances OsHKT1;5-Mediated Na+ Transport
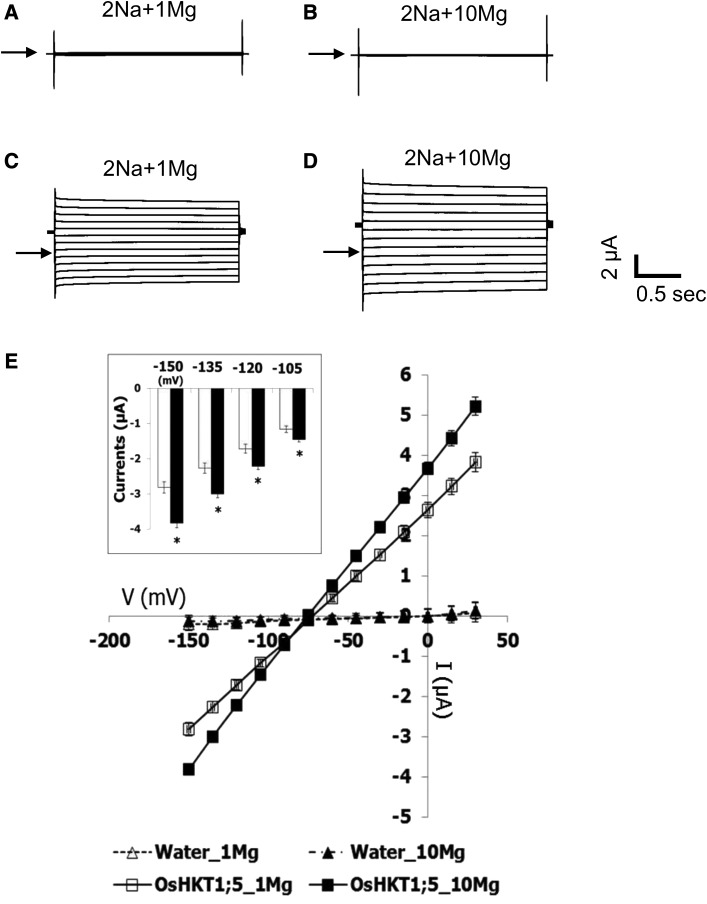
OsHKT1;5 is a key player for salt tolerance in rice (Ren et al., 2005). To link OsHKT1;5 and OsMGT1, we tested the effect of Mg2+ on OsHKT1;5-mediated Na+ transport in X. laevis oocytes by two-electrode voltage clamp (TEVC; Fig. 7). Water-injected control oocytes showed small background currents in the presence of 1 mm and 10 mm Mg2+ with 2 mm Na+, and no characteristic difference was found between the two Mg2+ concentrations tested (Fig. 7, A and B). In contrast, an increase of the Mg2+ concentration from 1 mm to 10 mm in the 2 mm Na+ bath solution elicited larger inward and outward currents from OsHKT1;5-expressing oocytes (Fig. 7, C and D). Significant increases in inward Na+ currents were found in OsHKT1;5-expressing oocytes at the membrane potentials of −105, −120, −135, and −150 mV upon the 10-fold Mg2+ increase in the bath (P < 0.05; Fig. 7E, inset). Current voltage relationships indicated that a change of Mg2+ concentration in the bath solution leads to no remarkable influence on the reversal potential of water-injected oocytes and OsHKT1;5-expressing oocytes (Fig. 7E). In addition, increasing Mg2+ concentration from 0.1 to 1 mm did not significantly increase inward and outward currents from OsHKT1;5-expressing oocytes (Supplemental Fig. S5). These results suggest that high Mg2+ concentration at millimolar level is required to enhance the OsHKT1;5-mediated Na+ transport.
Figure 7.
Enhancement of OsHKT1;5-mediated Na+ transport by Mg2+. TEVC experiments were performed using X. laevis oocytes injected with water or 12.5 ng of OsHKT1;5 cRNA in the presence of different concentrations of Mg2+. A, A current profile of water-injected oocytes, bathed in a 2 mm Na+ solution containing 1 mm MgCl2. B, A current profile of water-injected oocytes, bathed in a 2 mm Na+ solution containing 10 mm MgCl2. C, A current profile of OsHKT1;5-expressing oocytes, bathed in a 2 mm Na+ solution containing 1 mm MgCl2. D, A current profile of OsHKT1;5-expressing oocytes, bathed in a 2 mm Na+ solution containing 10 mm MgCl2. Arrowheads indicate zero current potentials in A to D. E, Current-voltage relationships of oocytes injected with water or 12.5 ng of OsHKT1;5 cRNA are shown (n = 6 for each; ±se): white triangle, water-injected oocytes in 1 mm Mg2+; black triangle, water-injected oocytes in 10 mm Mg2+; white square, OsHKT1;5-expressing oocytes in 1 mm Mg2+; black square, OsHKT1;5-expressing oocytes in 10 mm Mg2+. Voltage steps from +30 to −150 mV were applied with a holding potential of −40 mV. An inset indicates OsHKT1;5-mediated inward currents at −105, −120, −135, and −150 mV in the presence of 1 mm and 10 mm MgCl2 (open bars and closed bars, respectively). Asterisks in the inset represent a significant difference between 1 and 10 mm Mg2+ conditions at each membrane potential (P < 0.05, the Student’s t test).
We also performed the TEVC experiment in oocytes to investigate whether OsHKT1;5 can be enhanced by Ca2+. The result showed that under high concentration of Ca2+ (18 mm), the inward and outward current generated from OsHKT1;5-expressing oocytes was not significantly enhanced when compared with low concentration of Ca2+ (1.8 mm) (Supplemental Fig. S6), suggesting that OsHKT1;5 could not be enhanced by Ca2+.
OsHKT1;5 Knockout Line Shows Similar Phenotype to OsMGT1 Knockout Line under Salt Stress
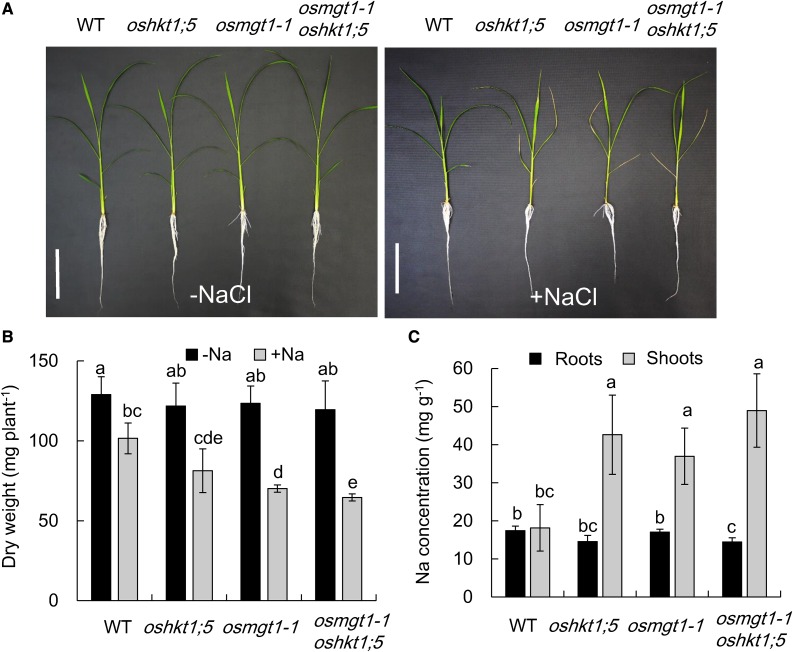
To further confirm the relationship between OsHKT1;5 and OsMGT1, we compared the phenotype of single knockout mutant of OsHKT1;5 and OsMGT1, and a double mutant of OsHKT1;5 and OsMGT1 under salt stress. There was no difference in the growth among wild type, single, and double mutants in the absence of NaCl (Fig. 8, A and B). However, in the presence of NaCl, the lower leaves of single and double mutants were rolled and wilt. The dry weight was decreased by about 41% by NaCl in the single and double mutants in contrast to 21% decrease in wild type (Fig. 8B). The Na concentration in the shoots of single and double mutants was about 2-fold of wild type (Fig. 8C). There were no differences in the dry weight and Na accumulation in the shoots between single and double mutants (Fig. 8, B and C). We further investigated whether high Mg2+ can alleviate salt stress-induced growth inhibition in oshkt1;5 and the double mutant (Supplemental Fig. S7). Different from osmgt1 mutants, high Mg supply did not alter the salt sensitivity in oshkt1;5 and the double mutant (Supplemental Fig. S7), suggesting that Mg2+ restoration in rice salt tolerance is specific to OsMGT1 function.
Figure 8.
Sensitivity of osmgt1-1 oshkt1;5 double-knockout mutant to salt stress. A, Effect of salt stress on the growth of osmgt1-1 oshkt1;5 double mutants. Left, 0 mm NaCl; right, 50 mm NaCl. Bar = 10 cm. B, Dry weight of seedlings without or with exposure to salt stress. C, Na concentration in the roots and shoots. Seedlings of wild type, single osmgt1, and oshkt1;5 mutants and double mutant were exposed to a nutrient solution containing 0 or 50 mm NaCl for 7 d. Data are means ± sd (n = 3). The asterisk shows a significant difference compared with wild type (P < 0.05 by Tukey’s test).
DISCUSSION
Different Roles of OsMGT1 in Al Tolerance and Salt Tolerance
OsMGT1 is a plasma membrane-localized Mg2+ transporter and was found to be involved in Al tolerance in rice previously (Chen et al., 2012). Knockout of OsMGT1 resulted in decreased Mg in the roots and increased sensitivity to Al, although the shoot Mg concentration was not altered. In this study, we found that OsMGT1 was also involved in salt tolerance. In two independent knockout lines of OsMGT1, the sensitivity to salt stress was significantly increased (Fig. 1A). Especially, the lower leaves of the mutants were wilted due to Na+ toxicity. However, it seems that OsMGT1 plays different roles in Al tolerance and salt tolerance. The site of Al3+ toxicity, which is characterized by rapid root elongation inhibition, is located at the root tips (Ryan et al., 1993). Therefore, OsMGT1 expressed at the root tip is responsible for Al tolerance, which is supported by the expression pattern of OsMGT1. OsMGT1 is up-regulated by Al3+ in the root tips (Chen et al., 2012), and its expression is regulated by ART1, a C2H2 zinc-finger type transcription factor (Yamaji et al., 2009). Furthermore, it is localized to the all cells of the root tips (Fig. 6, A and B). This localization is similar to other ART1-regulated genes (Huang et al., 2009, 2012; Xia et al., 2010, 2013; Yokosho et al., 2011; Che et al., 2016). Therefore, Al3+-induced up-regulation of OsMGT1 in the root tips results in increased cytosolic Mg2+ concentration, which prevents Al3+ binding to oxygen donor compounds because Al3+ and Mg2+ ions have similar hydrated radius (Bose et al., 2011).
By contrast, OsMGT1 expressed in the root mature region is responsible for salt tolerance. The root mature region has a developed vascular system and is the zone responsible for the root-to-shoot translocation of Na. The expression of OsMGT1 in the root mature zone was up-regulated by salt stress, although the induction was not as large as that by Al3+ stress (Fig. 5; Chen et al., 2012). Furthermore, different from the root tips, OsMGT1 in the root mature zone was highly expressed in the stellar tissues, especially in the root pericycle, which connects to the xylem and phloem (Fig. 6, C and D). Knockout of OsMGT1 resulted in significant high root-to-shoot translocation of Na, leading to the accumulation of Na in the leaves (Figs. 2B and 3A). These findings indicate that OsMGT1 in the root mature zone is involved in restriction of Na to the shoots. Therefore, the physiological role of OsMGT1 depends on its spatial expression in the roots.
OsMGT1 Confers Salt Tolerance through OsHKT1;5-Mediated Xylem Na+ Unloading in Roots
Several salt tolerance genes in rice have been reported including OsSOS1, OsHKT1;1, OsHKT1;4, and OsHKT1;5, which function as Na+ transporters (Martínez-Atienza et al., 2007; Wang et al., 2015; Suzuki et al., 2016; Ren et al., 2005). There is a possibility that Mg2+ transported by OsMGT1 regulates the transport activity of these transporters. OsSOS1 as a Na+/H+ antiporter was proposed to be responsible for extruding Na+ from the roots (Martínez-Atienza et al., 2007). The expression of OsSOS1 in the roots was not induced by NaCl within 24 h, and there was also no difference in the expression level of OsSOS1 between wild type and two osmgt1 mutants (Fig. 5G; Supplemental Fig. S4C). Since there was no difference in the total Na uptake between wild type and two osmgt1 mutants (Fig. 2A), it is unlikely that OsSOS1 activity is associated with OsMGT1. On the other hand, OsHKT1;1 was mainly expressed in the shoots (Wang et al., 2015), and the contribution of OsHKT1;4 to the Na exclusion from shoots was found to be low at the vegetative growth stage (Suzuki et al., 2016). Therefore, the possibility of these transporters in restricting Na accumulation through OsMGT1 is also excluded at the young seedling stage. By contrast, our results showed that the activity of OsHKT1;5 requires cooperation of OsMGT1. This is supported by several pieces of direct and indirect evidence. First, the activity of OsHKT1;5 was significantly enhanced by Mg2+ in TEVC experiments (Fig. 7). Second, increased salt sensitivity and Na accumulation in the shoots of osmgt1 mutants were completely restored at high Mg2+ supply (Fig. 4). Third, OsMGT1 expression showed similar tissue localization at the root stele as OsHKT1;5 in the root mature zones (Ren et al., 2005; Fig. 6, C and D). Fourth, knockout of OsHKT1;5 or OsMGT1 resulted in increased Na accumulation in the shoots similarly (Fig. 8C). Fifth, knockout of OsHKT1;5 in the background of osmgt1 mutant did not further increase salt tolerance and Na accumulation in the shoots (Fig. 8). OsHKT1;5 has been strongly suggested to play a key function in restricting Na accumulation in the shoots by excluding Na+ from the xylem, and its differential expression is also responsible for genotypic difference in salt tolerance of rice (Ren et al., 2005; Cotsaftis et al., 2012; Deinlein et al., 2014). However, the expression of OsHKT1;5 did not change in osmgt1 mutants (Supplemental Fig. S4B). Our results therefore indicate that Mg2+ transported by OsMGT1 is required for enhancing OsHKT1;5 activity in the stellar tissues of root mature zone, conferring salt tolerance in rice. The exact mechanism underlying this activation is unknown, but the possible mode is that Mg2+ is required for the stereostructure for OsHKT1;5 protein.
In addition to expression of OsMGT1 in the roots, its expression was also detected in the shoots, although it was not induced by salt stress (Fig. 5A). Furthermore, OsMGT1 was expressed in the phloem region of basal node, leaf blade, and sheath (Fig. 6, H–K). This tissue specificity of expression of OsMGT1 is similar to OsHKT1;1 and OsHKT1;4 (Wang et al., 2015; Suzuki et al., 2016). Therefore, there is a possibility that Mg2+ transported by OsMGT1 in these tissues also plays a role in enhancing the transport activity of OsHKT1;1 and OsHKT1;5, although further work remains to be done.
In conclusion, our results showed that OsMGT1 is involved in salt tolerance in rice by enhancing at least OsHKT1;5 activity. Mg2+ transported by OsMGT1 in the stellar tissues of the root mature zone enhances the Na+ transport activity of OsHKT1;5, resulting in decreased Na accumulation to the shoots and subsequently contributing to increased salt tolerance in rice.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Two Tos-17 insertion lines, NF0595 (osmgt1-1) and NE4528 (osmgt1-2) for OsMGT1, were obtained from the Rice Genome Resource Center in Japan. The homozygous insertion lines were isolated by PCR according to Chen et al. (2012). The oshkt1;5 mutant was a T-DNA insertion line (Supplemental Fig. S8A). RT-PCR was performed using gene-specific primers (RT-F, 5′-TGATGTACCTGCCACCTTAC-3′, and RT-R, 5′-ATGTTGAGGACGCTGTAGTT-3′) to genotype oshkt1;5 homozygous lines. Actin gene (5′-TTGGATTCTGGTGATGGTGT-3′ and 5′-GGAAGCTCGTAGCTCTTCTC-3′) was used as an internal control. No OsHKT1;5 transcript was present in this line (Supplemental Fig. S8B).
osmgt1 oshkt1;5 double-knockout mutants were generated by crossing homozygous osmgt1-1 (NF0595, Nipponbare background) with oshkt1;5 (Dongjing background). F1 heterozygote and F2 homozygous mutant plants were isolated by PCR genotyping (Supplemental Fig. S8C). The osmgt1 homozygous line was screened by PCR using specific primers (OsMGT1-F, 5′-AACACGCATCTAAAAGTTTCACC-3′ and OsMGT1-R, 5′-TTCGATTATTATTGCTCCCACA-3′) with a Tos-17 primer (Tos17-F, 5′-ATTGTTAGGTTGCAAGTTAGTTAAGA-3′). The oshkt1;5 homozygous line was screened by PCR using specific primers (OsHKT1;5-F, 5′-GCTGTTGCTCTTGTTCACTT-3′, and OsHKT1;5-R, 5′-CACGAACATCAGTAGCCTCG-3′) with a T-DNA border primer (LB-F, 5′-CTCTAGAGTCCAGAATTCAGTAC-3′).
Seeds were soaked in deionized water at 30°C in the dark for 2 d, and then transferred to a net floating on a 0.5 mm CaCl2 solution in a 1.5-liter plastic container for 7 d. The seedlings were grown in a glasshouse under natural light at 25°C to 30°C using 0.5× Kimura B nutrient solution for various experiments. Na+ and Mg2+ were applied as NaCl and MgCl2·6H2O, respectively. The solution in each experiment was renewed every 2 d.
Phenotypic Analysis and Elemental Determination
To compare the sensitivity to salt, high Mg, and Cd toxicity, seedlings of both wild-type and two OsMGT1 knockout lines were exposed to a nutrient solution containing 50 mm NaCl, 10 mm MgCl2, or 5 µm CdCl2. After 12 d, individual plant was photographed, and leaves were separated and photographed. Both roots and shoots were sampled after the roots were washed with 5 mm CaCl2 for three times to remove the apoplastic ions.
To investigate the Na accumulation in shoots, rice seedlings were exposed to a nutrient solution containing 50 mm NaCl for 3 d. The leaf blades 3 to 7 (from old to young) were separately sampled. Leaves 1 and 2 were too small to be sampled. Xylem sap was collected from the wild-type and the knockout lines exposed to a nutrient solution containing 10 mm NaCl for 24 h. The shoots (2 cm above the roots) were excised with a razor, and then the xylem sap was collected with a micropipette for 1 h after decapitation of the shoots.
The alleviative effect of Mg2+ on salt stress was investigated by exposing wild type (Nipponbare and Dongjing), osmgt1 mutants, oshkt1;5 mutant, and double mutant osmgt1 oshkt1;5 to a nutrient solution containing 0.02, 0.2, or 2 mm MgCl2 in the presence of 50 mm NaCl for 7 to 12 d. Both roots and shoots were sampled for mineral analysis as described below.
To investigate the sensitivity of osmgt1 oshkt1;5 double-knockout mutant to salt stress, the F3 progeny of wild type, single osmgt1 and oshkt1;5 mutants, and double mutants showing the same genetic background were exposed to a nutrient solution containing 0 or 50 mm NaCl. After 7 d exposure, both the roots and shoots were sampled for analysis.
After harvest, the roots and shoots were dried at 70°C for 2 d and then were subjected to digestion with concentrated HNO3. The metal concentration in the digested solution and xylem sap was determined by inductively coupled plasma-mass spectrometry (ICP-MS 7700X; Agilent Technologies). The Na content was calculated based on Na concentration and dry weight.
RNA Isolation and Expression Analysis
For time-response expression analysis, 14-d-old rice seedlings (cv Nipponbare) were exposed to a 0.5× Kimura B solution containing 50 mm NaCl for different hours, and the roots and shoots were sampled for RNA extraction. For dose-dependent expression, rice seedlings were exposed to a 0.5× Kimura B solution containing different concentrations of NaCl for 6 h, and the roots were sampled for RNA extraction. For root spatial expression, different root segments (0–5, 5–10, and 10–20 mm from root tips) were excised after the roots exposed 0 or 50 mm NaCl for 24 h. To compare the salt tolerance genes expression between wild type and osmgt1 mutants, rice seedlings of both wild type and osmgt1 mutants were exposed to a 0.5× Kimura B solution with or without 50 mm of NaCl for 24 h, and the roots were sampled for RNA extraction.
Total RNA from rice roots and shoots was extracted using the RNeasy Mini Kit (Qiagen). One microgram of total RNA was used for first-strand cDNA synthesis using a ReverTra Ace qPCR RT Master Mix kit (TOYOBO) following the manufacturer’s instructions.
The gene expression level was determined by real-time RT-PCR using Thunderbird SYBR qPCR Mix (TOYOBO) on Mastercycler ep realplex (Eppendorf). The primers used were 5′-GGCGCGTGCAGAAGATTAGGG-3′ and 5′-CGCGTATTCACGGATATGGTACAGGG-3′ for OsMGT1; 5′-TCAATCCAGACCATCTCTGC-3′ and 5′-CTAAGTTTCCAGGCTTTGCC-3′ for OsHKT1;4; 5′-TGATGTACCTGCCACCTTAC-3′ and 5′-ATGTTGAGGACGCTGTAGTT-3′ for OsHKT1;5; 5′-TCTGAAACCAGTTGACGGAA-3′ and 5′-AGATCACCAAAAGCCTCCAA-3′ for OsSOS1. Actin was used as an internal control. Normalized relative expression was calculated by the ΔΔCt method.
Tissue Specificity of Expression
The native 2.5-kb promoter sequence of OsMGT1 was linked to sGFP gene by overlap PCR. The amplified PCR product was inserted into pPZP2H-lac (with NOS terminator) using ApaI and SpeI to create the OsMGT1 promoter-GFP construct. The construct was introduced into the calluses of rice (cv Nipponbare) via Agrobacterium tumefaciens-mediated transformation (Hiei et al., 1994). The primer sequences (5′-AATGGGCCCatgcccatcaacgacgacat-3′ and 5′-CGGACTAGTGCCGCTTTACTTGTACAG-3′) were used for amplification and introduction of the ApaI and SpeI restriction sites. Different organs and tissues were sampled for the immunostaining using an antibody against GFP (A11122; Molecular Probes) as described previously (Yamaji and Ma, 2007). For NaCl-responsive assay, root segments (20 mm from root tips) were sampled after the roots exposed to 0 or 50 mm NaCl for 24 h. Fluorescence of the secondary antibody (Alexa Fluor 555 goat anti-rabbit IgG; Molecular Probes) was observed with a confocal laser-scanning microscope (LSM700; Carl Zeiss).
Na Accumulation and Distribution in the Shoots
Seedlings (14 d old) of both wild-type rice and two knockout lines were treated with 50 mm Na for 7 d. Shoot basal region about 20 mm above the root-shoot junction was excised and embedded in 5% agar. The shoot transverse section at 10 mm from the basal region was cut and immediately used for analysis by scanning electron microscope (TM3000; Hitachi) in vacuum condition at −20°C. The elemental distribution photos were generated by energy dispersive x-ray spectrometer (SwiftED 3000; Oxford Instruments).
OsHKT1;5 Expression Construct and TEVC Experiments Using Xenopus laevis Oocytes
The coding region of OsHKT1;5 cDNA was isolated from a japonica rice cultivar Nipponbare based on the sequence information at GenBank. The isolated cDNA was further amplified by the primers harboring the BglII site: Fw, 5′-AAGATCTATGAGTTCTCTGGATGCCACTA-3′ and Rev, 5′-CATGAGATCTTTATTCTATCTTCCATGCCTGACCA-3′ using the Phusion high-fidelity DNA polymerase (New England Biolabs). The sequence of the amplified fragment was checked and then subcloned at the BglII site of the pXβG-ev1 vector (Suzuki et al., 2016). OsHKT1;5 cRNA was transcribed using a mMESSAGE mMACHINE in vitro transcription kit (Ambion). TEVC experiments using oocytes of X. laevis frogs were performed as described previously (Suzuki et al., 2016). In brief, water or 12.5 ng of OsHKT1;5 cRNA was injected into oocytes and incubated at 18°C for 1 d. TEVC experiments were performed using an Axoclamp 900A amplifier (Molecular Devices) and an Axon Instruments Digidata 1440A and pCLAMP 10 (Molecular Devices). Oocytes were bathed in a solution containing 2 mm Na-Glu, 1.8 mm or 18 mm CaCl2, 10 mm MES-1,3-bis (tris(hydroxymethyl]methylamino) propane, 180 mm d-mannitol (pH 5.5; with BisTrisPropane), and 0.1 mM, 1 mm, or 10 mm MgCl2. The osmolality of each solution was adjusted to 220 to 240 mosmol kg−1 with d-mannitol. Voltage steps were applied from +30 to −120 or −150 mV in 15-mV decrements, with a holding potential of −40 mV. All experiments were performed at room temperature.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AB731703 for OsMGT1 and AK108663 for SKC1/OsHKT1;5.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Sensitivity of OsMGT1 knockout lines to osmotic stress and water deficit.
Supplemental Figure S2. Sensitivity of OsMGT1 knockout lines to Mg and Cd toxicity
Supplemental Figure S3. Comparison of Mg and K in the shoots, roots, and xylem sap of wild-type rice and knockout lines of OsMGT1.
Supplemental Figure S4. Gene expression of OsHKT1;4, OsHKT1;5, and OsSOS1 in wild-type and OsMGT1 knockout lines.
Supplemental Figure S5. Regulation of OsHKT1;5-mediated Na+ transport by 0.1 or 1.0 mm Mg2+.
Supplemental Figure S6. Regulation of OsHKT1;5-mediated Na+ transport by 1.8 or 18 mm Ca2+.
Supplemental Figure S7. Effect of high Mg2+ supply on salt sensitivity in oshkt1;5 and osmgt1-1 oshkt1;5 double mutant.
Supplemental Figure S8. Isolation of osmgt1-1 oshkt1;5 double-knockout rice mutants.
Acknowledgments
We thank the Rice Genome Resource Center in Tsukuba for providing the Tos-17 insertion line, and Maki Katsuhara and Osamu Shimotani for the preparation of oocytes. We also thank Nao Komiyama help generating transgenic rice.
Glossary
- TEVC
two-electrode voltage clamp
Footnotes
This work was supported by a Grant-in-Aid for Specially Promoted Research (JSPS KAKENHI grant number 16H06296 to J.F.M.) and by the Ministry of Education, Culture, Sports, Science and Technology as part of the Joint Research Program implemented at the Institute of Plant Science and Resources, Okayama University in Japan (2615, 2716 to T.H.), and by the National Natural Science Foundation of China (No. 31672218 to Z.C.C.).
Articles can be viewed without a subscription.
References
- Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285: 1256–1258 [DOI] [PubMed] [Google Scholar]
- Apse MP, Sottosanto JB, Blumwald E (2003) Vacuolar cation/H+ exchange, ion homeostasis, and leaf development are altered in a T-DNA insertional mutant of AtNHX1, the Arabidopsis vacuolar Na+/H+ antiporter. Plant J 36: 229–239 [DOI] [PubMed] [Google Scholar]
- Bose J, Babourina O, Rengel Z (2011) Role of magnesium in alleviation of aluminum toxicity in plants. J Exp Bot 62: 2251–2264 [DOI] [PubMed] [Google Scholar]
- Byrt CS, Xu B, Krishnan M, Lightfoot DJ, Athman A, Jacobs AK, Watson-Haigh NS, Plett D, Munns R, Tester M, et al. (2014) The Na+ transporter, TaHKT1;5-D, limits shoot Na+ accumulation in bread wheat. Plant J 80: 516–526 [DOI] [PubMed] [Google Scholar]
- Che J, Yamaji N, Shen RF, Ma JF (2016) An Al-inducible expansin gene, OsEXPA10 is involved in root cell elongation of rice. Plant J 88: 132–142 [DOI] [PubMed] [Google Scholar]
- Chen H, An R, Tang JH, Cui XH, Hao FS, Chen J, Wang XC (2007) Over-expression of a vacuolar Na+/H+ antiporter gene improves salt tolerance in an upland rice. Mol Breed 19: 215–225 [Google Scholar]
- Chen ZC, Ma JF (2013) Magnesium transporters and their role in Al tolerance in plants. Plant Soil 368: 51–56 [Google Scholar]
- Chen ZC, Yamaji N, Motoyama R, Nagamura Y, Ma JF (2012) Up-regulation of a magnesium transporter gene OsMGT1 is required for conferring aluminum tolerance in rice. Plant Physiol 159: 1624–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotsaftis O, Plett D, Shirley N, Tester M, Hrmova M (2012) A two-staged model of Na+ exclusion in rice explained by 3D modeling of HKT transporters and alternative splicing. PLoS One 7: e39865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport R, James RA, Zakrisson-Plogander A, Tester M, Munns R (2005) Control of sodium transport in durum wheat. Plant Physiol 137: 807–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deinlein U, Stephan AB, Horie T, Luo W, Xu GH, Schroeder JI (2014) Plant salt-tolerance mechanisms. Trends Plant Sci 19: 371–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert M, Meschenmoser K, Svidova S, Weghuber J, Schweyen R, Eifler K, Lenz H, Weyand K, Knoop V (2009) A root expressed magnesium transporter of the MRS2/MGT gene family in Arabidopsis thaliana allows for growth in low-Mg environments. Plant Cell 21: 4018–4030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamoto S, Horie T, Hauser F, Deinlein U, Schroeder JI, Uozumi N (2015) HKT transporters mediate salt stress resistance in plants: from structure and function to the field. Curr Opin Biotechnol 32: 113–120 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Hmiel SP, Snavely MD, Miller CG, Maguire ME (1986) Magnesium transport in Salmonella typhimurium: Characterisation of magnesium influx and cloning of a transport gene. J Bacteriol 168: 1444–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Karahara I, Katsuhara M (2012) Salinity tolerance mechanisms in glycophytes: An overview with the central focus on rice plants. Rice (NY) 5: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CF, Yamaji N, Chen Z, Ma JF (2012) A tonoplast-localized half-size ABC transporter is required for internal detoxification of aluminum in rice. Plant J 69: 857–867 [DOI] [PubMed] [Google Scholar]
- Huang CF, Yamaji N, Mitani N, Yano M, Nagamura Y, Ma JF (2009) A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell 21: 655–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabnoune M, Espeout S, Mieulet D, Fizames C, Verdeil JL, Conéjéro G, Rodríguez-Navarro A, Sentenac H, Guiderdoni E, Abdelly C, et al. (2009) Diversity in expression patterns and functional properties in the rice HKT transporter family. Plant Physiol 150: 1955–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James RA, Blake C, Byrt CS, Munns R (2011) Major genes for Na+ exclusion, Nax1 and Nax2 (wheat HKT1;4 and HKT1;5), decrease Na+ accumulation in bread wheat leaves under saline and waterlogged conditions. J Exp Bot 62: 2939–2947 [DOI] [PubMed] [Google Scholar]
- James RA, Davenport RJ, Munns R (2006) Physiological characterization of two genes for Na+ exclusion in durum wheat, Nax1 and Nax2. Plant Physiol 142: 1537–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidi EO, Barragán V, Rubio L, El-Hamdaoui A, Ruiz MT, Cubero B, Fernández J, Bressan RA, Hasegawa PM, Quintero FJ, et al. (2010) The AtNHX1 exchanger mediates potassium compartmentation in vacuoles of transgenic tomato. Plant J 61: 495–506 [DOI] [PubMed] [Google Scholar]
- Li L, Tutone AF, Drummond RS, Gardner RC, Luan S (2001) A novel family of magnesium transport genes in Arabidopsis. Plant Cell 13: 2761–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutts S, Kinet JM, Bouharmont J (1996) Effects of salt stress on growth, mineral nutrition and proline accumulation in relation to osmotic adjustment in rice (Oryza sativa L.) cultivars differing in salinity resistance. Plant Growth Regul 19: 207–218 [Google Scholar]
- Mao D, Chen J, Tian L, Liu Z, Yang L, Tang R, Li J, Lu C, Yang Y, Shi J, et al. (2014) Arabidopsis transporter MGT6 mediates magnesium uptake and is required for growth under magnesium limitation. Plant Cell 26: 2234–2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Atienza J, Jiang X, Garciadeblas B, Mendoza I, Zhu JK, Pardo JM, Quintero FJ (2007) Conservation of the salt overly sensitive pathway in rice. Plant Physiol 143: 1001–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäser P, Eckelman B, Vaidyanathan R, Horie T, Fairbairn DJ, Kubo M, Yamagami M, Yamaguchi K, Nishimura M, Uozumi N, et al. (2002) Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett 531: 157–161 [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX (2005) A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet 37: 1141–1146 [DOI] [PubMed] [Google Scholar]
- Rosas-Santiago P, Lagunas-Gómez D, Barkla BJ, Vera-Estrella R, Lalonde S, Jones A, Frommer WB, Zimmermannova O, Sychrová H, Pantoja O (2015) Identification of rice cornichon as a possible cargo receptor for the Golgi-localized sodium transporter OsHKT1;3. J Exp Bot 66: 2733–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan PR, Ditomaso JM, Kochian LV (1993) Aluminium toxicity in roots: An investigation of spatial sensitivity and the role of the root cap. J Exp Bot 44: 437–446 [Google Scholar]
- Schock I, Gregan J, Steinhauser S, Schweyen R, Brennicke A, Knoop V (2000) A member of a novel Arabidopsis thaliana gene family of candidate Mg ion transporters complements a yeast mitochondrial group II intron-splicing mutant. Plant J 24: 489–501 [DOI] [PubMed] [Google Scholar]
- Shabala S, Cuin TA (2008) Potassium transport and plant salt tolerance. Physiol Plant 133: 651–669 [DOI] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97: 6896–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunarpi, Horie T, Motoda J, Kubo M, Yang H, Yoda K, Horie R, Chan WY, Leung HY, Hattori K, et al. (2005) Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na+ unloading from xylem vessels to xylem parenchyma cells. Plant J 44: 928–938 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Yamaji N, Costa A, Okuma E, Kobayashi NI, Kashiwagi T, Katsuhara M, Wang C, Tanoi K, Murata Y, et al. (2016) OsHKT1;4-mediated Na+ transport in stems contributes to Na+ exclusion from leaf blades of rice at the reproductive growth stage upon slat stress. BMC Plant Biol 16: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Jing W, Xiao L, Jin Y, Shen L, Zhang W (2015) The OsHKT1;1 transporter is involved in salt tolerance and regulated by an MYB-type transcription factor. Plant Physiol 168: 1076–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Yamaji N, Kasai T, Ma JF (2010) Plasma membrane-localized transporter for aluminum in rice. Proc Natl Acad Sci USA 107: 18381–18385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Yamaji N, Ma JF (2013) A plasma membrane-localized small peptide is involved in rice Al tolerance. Plant J 76: 345–355 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Hamamoto S, Uozumi N (2013) Sodium transport system in plant cells. Front Plant Sci 4: 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji N, Huang CF, Nagao S, Yano M, Sato Y, Nagamura Y, Ma JF (2009) A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell 21: 3339–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji N, Ma JF (2007) Spatial distribution and temporal variation of the rice silicon transporter Lsi1. Plant Physiol 143: 1306–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokosho K, Yamaji N, Ma JF (2011) An Al-inducible MATE gene is involved in external detoxification of Al in rice. Plant J 68: 1061–1069 [DOI] [PubMed] [Google Scholar]
- Zhu JK. (2001) Plant salt tolerance. Trends Plant Sci 6: 66–71 [DOI] [PubMed] [Google Scholar]