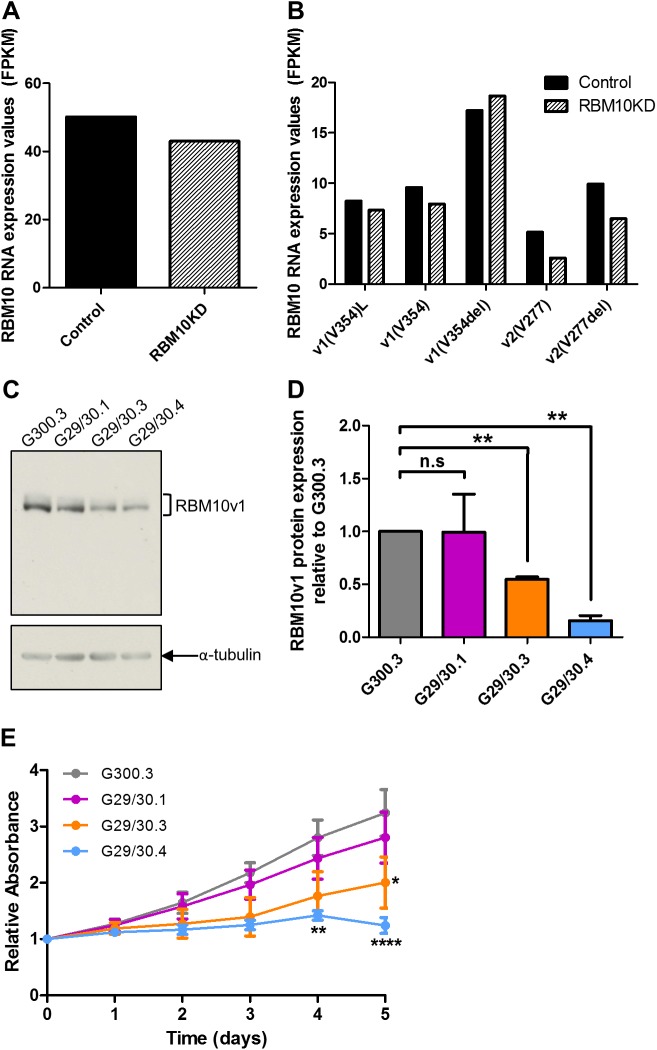
Fig 3. RBM10 expression and dehydrogenase activity in control and stable RBM10KD GLC20 sublines.
(A & B) RBM10 mRNA levels, determined by RNA-Seq, for all RBM10 variants combined (A) or for individual splice variants (B). ‘Control’ represents values from parental GLC20 cells and the control G300.3 GLC20 subline. RBM10KD sample used was the G29/30.4 subline, as it showed greatest decrease in RBM10 protein expression. (C & D) RBM10v1 protein levels were monitored in the G300.3, G29/30.1, G29/30.3 and G29/30.4 sublines using the Bethyl RBM10 antibody. (C) One representative Western blot result is presented for RBM10v1 and α-tubulin (loading control). (D) Densitometric results of the average RBM10v1 protein levels from three biological replicates performed in duplicate. Analysis was performed using the AlphaEaseFC, ‘1D-Multi’ analysis tool. Values of RBM10v1 were normalized to the α-tubulin of each biological replicate, and then made relative to the G300.3 control subline. Standard error is presented. Subline expression levels were compared using the Student’s unpaired t-test, between sublines. (E) G300.3, G29/30.1, G29/30.3 and G29/30.4 were grown for five days and dehydrogenase activity was monitored daily using an MTT assay. Absorbance was plotted relative to day 0. The average of three biological replicates performed in eight technical replicates with standard error is presented. A Two-way ANOVA was performed between the G300.3 and other sublines, with Bonferroni post-hoc analysis. * p < 0.05, ** p < 0.01, and **** p < 0.0001.