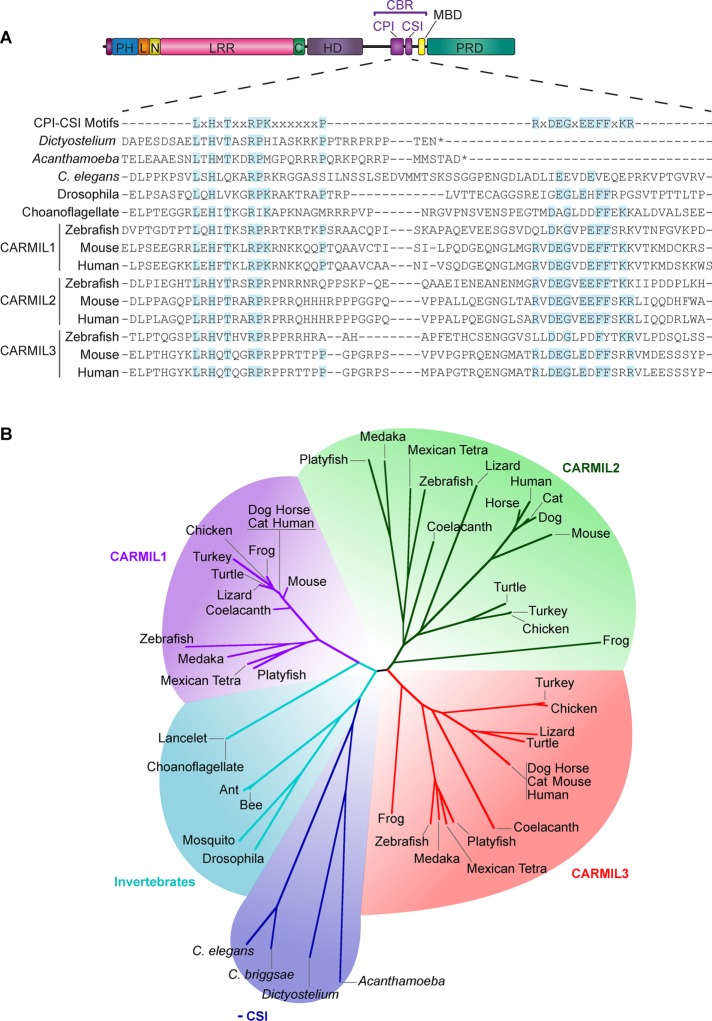
FIGURE 1:
Conservation and domain architecture of CARMIL proteins. (A) Domain architecture of CARMIL proteins, illustrating the arrangement of the PH domain, linker (L), N-cap (N), LRR domain, C-cap (C), HD, CBR consisting of a CPI motif and a CSI motif, MBD, and a PRD. Sequence alignment of the CBR for selected CARMILs, including the three vertebrate isoforms in zebrafish, mouse, and human (encoded by three separate genes). The alignment includes sequences for invertebrates, which lack the CSI motif region or lack residues known to be required for the CSI-CP interaction (Zwolak et al., 2010b). Shaded residues are identical to the consensus sequences. (B) Unrooted phylogenetic tree showing the relationships among CARMIL proteins, revealing five groups of CARMIL genes. Vertebrate genomes have three genes that encode three conserved isoforms—CARMIL1, CARMIL2, and CARMIL3—and invertebrates have a single CARMIL gene and isoform. Invertebrate CARMILs can be further classified into those that contain a CSI motif and those that do not.