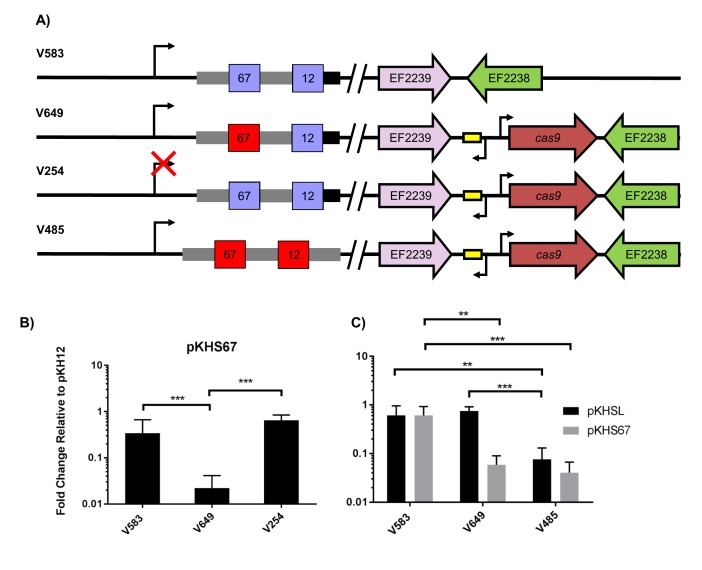
Figure 2. Assessment of CRISPR2 function in V583.
(A) Genotypes of relevant strains are diagrammed. In V485, the CRISPR2 terminal repeat is replaced with a direct repeat. Spacers in red were experimentally determined to provide genome defense, while blue spacers do not provide genome defense. (B) Conjugation frequencies relative to pKH12 are plotted for pKHS67 transfer into V583, V649, and V254. The geometric mean and geometric standard deviation are shown for 4 independent biological replicates. (C) Conjugation frequencies relative to pKH12 are plotted for pKHSL and pKHS67 transfer into V583, V649, and V485. pKHSL contains the S12 sequence from the V583 CRISPR2 array. The geometric mean and geometric standard deviation for 3 independent biological replicates are shown. ***p<0.001, **p<0.01.