Abstract
Background
Amphimeriasis is a fish-borne disease caused by the liver fluke Amphimerus spp. that has recently been reported as endemic in the tropical Pacific side of Ecuador with a high prevalence in humans and domestic animals. The diagnosis is based on the stool examination to identify parasite eggs, but it lacks sensitivity. Additionally, the morphology of the eggs may be confounded with other liver and intestinal flukes. No immunological or molecular methods have been developed to date. New diagnostic techniques for specific and sensitive detection of Amphimerus spp. DNA in clinical samples are needed.
Methodology/Principal findings
A LAMP targeting a sequence of the Amphimerus sp. internal transcribed spacer 2 region was designed. Amphimerus sp. DNA was obtained from adult worms recovered from animals and used to optimize the molecular assays. Conventional PCR was performed using outer primers F3-B3 to verify the proper amplification of the Amphimerus sp. DNA target sequence. LAMP was optimized using different reaction mixtures and temperatures, and it was finally set up as LAMPhimerus. The specificity and sensitivity of both PCR and LAMP were evaluated. The detection limit was 1 pg of genomic DNA. Field testing was done using 44 human stool samples collected from localities where fluke is endemic. Twenty-five samples were microscopy positive for Amphimerus sp. eggs detection. In molecular testing, PCR F3-B3 was ineffective when DNA from fecal samples was used. When testing all human stool samples included in our study, the diagnostic parameters for the sensitivity and specificity were calculated for our LAMPhimerus assay, which were 76.67% and 80.77%, respectively.
Conclusions/Significance
We have developed and evaluated, for the first time, a specific and sensitive LAMP assay for detecting Amphimerus sp. in human stool samples. The procedure has been named LAMPhimerus method and has the potential to be adapted for field diagnosis and disease surveillance in amphimeriasis-endemic areas. Future large-scale studies will assess the applicability of this novel LAMP assay.
Author summary
Amphimeriasis, a fish-borne zoonotic disease caused by the liver fluke Amphimerus spp., is a highly prevalent parasitic infection affecting an indigenous Amerindian group, the Chachi, living in rural and remote tropical areas along the Río Cayapas and its tributaries in the north-western coastal rainforest of Ecuador. Very little is known about the clinical course and treatment of this disease, and the only method for diagnosing it is the parasitological microscopic detection of eggs from Amphimerus spp. in patients' stool samples. This method lacks sensitivity, and the morphology of the eggs may be confounded with other liver and intestinal flukes. New diagnostic tools that can improve the sensitivity and specificity for diagnosing Amphimerus spp. infection would be desirable. At present, LAMP technology shows all the characteristics required of a real-time assay with simple operation for potential use in the clinical diagnosis of infectious diseases, particularly in the field conditions in developing countries for most neglected tropical diseases. In this study, we developed and successfully evaluated a LAMP assay for detecting Amphimerus sp. in human stool samples. After further validation, our LAMP assay (LAMPhimerus) could be readily adapted for effective field diagnosis and disease surveillance in amphimeriasis-endemic areas.
Introduction
Amphimerus spp. are digenean parasitic flatworms in the bile ducts of birds, reptiles and mammals, and they are closely related to the genera Clonorchis and Opisthorchis within the Opisthorchiidae family [1, 2]. As for other members of the Opisthorchiidae family, the life cycle of Amphimerus spp. is highly complex, involving both freshwater snails and fish as intermediate hosts and vertebrates, including humans, as definitive hosts [3]. Humans or fish-eating animals are infected with Amphimerus spp. through the ingestion of raw or undercooked freshwater fish containing metacercariae [3]. Recently, Amphimerus sp. has been reported, for the first time, as endemic in rural communities in the tropical Pacific side of Ecuador with a high prevalence in humans and domestic cats and dogs, causing amphimeriasis [3, 4]. Several foodborne trematodiases around the world are now considered by the World Health Organization as neglected tropical diseases (NTDs) [5] with high prevalence, especially in East Asia [6], and they have serious consequences, such as cholangiocarcinoma [7,8]. Amphimeriasis has been reported as a new emerging foodborne zoonotic disease [3].
Amphimerus spp. adult stages are located in the bile ducts of the definitive host, and the eggs are shed in the feces [3]. Diagnosis of human and animal infection can be performed with the wet mount technique for examining feces, allowing for microscopic visualization of parasite eggs; the formalin-ether concentration method has been shown to increase the sensitivity ten-fold [3]. Detection of the eggs in bile or duodenal fluid can also be performed. However, microscopic examination is cumbersome and time consuming, and it could have a low sensitivity in cases of light infections. In addition, the morphological similarity of the Amphimerus spp. eggs to those of closely related species belonging to genera Clonorchis and Opisthorchis as well as to minute intestinal flukes, makes diagnosis difficult. It would be necessary to use scanning electron microscopy to accurately observe the differences between the coatings of the different species [3]. Therefore, the development of a new method that can improve the sensitivity and specificity for diagnosing Amphimerus spp. infection is urgently required.
To overcome these limitations, the use of molecular approaches has become a powerful tool for the diagnosis, identification and differentiation of closely related species. In recent years, several polymerase chain reaction (PCR)-based molecular diagnostic methods have been developed for detecting many parasitic trematodes, including those species that are closely related to Amphimerus spp., such as C. sinensis [9–14] and O. viverrini [15–18]. Although these studies have demonstrated that PCR-based methods are very sensitive and specific, they are not still widely used in low-income countries because well-trained personnel and expensive equipment are needed, making them unviable for routine application in field conditions in endemic areas that are generally undeveloped and have a high disease prevalence. Loop-mediated isothermal amplification (LAMP) could be a good alternative amplification technology [19] because it has several salient advantages over most PCR-based methods [20, 21]. At present, LAMP technology has all the characteristics required of a real-time assay along with simple operation for potential use in the clinical diagnosis of infectious diseases, particularly under the field conditions in developing countries [22, 23]. Additionally, several LAMP assays have already been successfully described for detecting trematode parasites, including a number of species causing foodborne trematodiases, such as Fasciola spp. [24], Clonorchis sinensis [25, 26], Opisthorchis viverrini [27–29] and Paragonimus westermani [30].
With the aim of developing new, applicable and cost-effective molecular tools for the diagnosis of amphimeriasis, we have developed and evaluated, for the first time, a LAMP assay for the specific detection of Amphimerus sp. liver fluke in human stool samples.
Methods
Ethics statement
The study protocol was approved by the Ethics Committee of Universidad Central del Ecuador (License number: LEC IORG 0001932, FWA 2482, IRB 2483. COBI-AMPHI-0064-11) and the Ethics Committee of the University of Salamanca (protocol approval number 48531). Participants were given detailed explanations about the aims, procedures and possible benefits of the study. Written informed consent was obtained from all subjects prior to the collection of biological samples for parasitological and molecular evaluation. Parents or guardians of children who participated in the study provided written informed consent on the child's behalf. All samples were coded and treated anonymously.
Study area and population
The study was conducted during February 2016 in two indigenous Chachi villages alongside the Cayapas River in the Esmeraldas province, located in the northwest coastal rainforest of Ecuador [4]. The indigenous Chachi, living together with the Afro-ecuadorian and mestizo populations, belong to the predominant autochthonous group in this area, representing 13% of the inhabitants in this region. These communities are the same as those studied previously and have a high prevalence of infection (15.5% to 34.1%) with Amphimerus sp. Prevalences are also high in local cats and dogs [3, 4]. They live in remote villages where the only way to reach them is by boat along the river. Sanitation facilities are lacking, and the members are hunters who habitually eat undercooked freshwater fish (mainly smoked fish) caught in the neighboring rivers [4]. More details on the region can be accessed elsewhere [31, 32].
Human stool samples and parasitological tests
Human stool samples were obtained from indigenous Chachi communities during February 2016. Each participant who enrolled in the study was given a copro-parasitological flask for stool collection. Samples were collected within a few hours of stool passing. After collection, samples were transported to the Parasitology Laboratory (Centro de Biomedicina, Universidad Central del Ecuador, Quito, Ecuador) for parasitological screening under light microscopy by direct examination, simple sedimentation, formalin-ether concentration and Kato-Katz techniques. All samples were examined by two qualified laboratory technicians according to the basic laboratory methods in medical parasitology recommended by the World Health Organization (WHO) [33]. After parasitological screening, a total of 44 stool samples were selected, including 25 (56.81%) that were positive for Amphimerus sp. eggs-by one or more parasitological methods-and 19 (43.18%) negative samples. Afterwards, the 44 stool samples that were well-preserved in 80% ethanol were sent to the Research Center for Tropical Diseases (CIETUS) at the University of Salamanca, Spain, for further DNA extraction and molecular analysis as described below.
DNA extraction for molecular analyses
DNA from human fecal samples
Approximately 250–300 mg from each of 44 stool samples preserved in 80% ethanol solution was used for DNA extraction. First, excess ethanol was removed from each vial; subsequently, DNA extraction was performed using the Mini Stool DNA Extraction kit (Macharey-Nagel) according to the manufacturers’ instructions. Purified DNA samples were stored at -20°C until use.
DNA from parasites
Amphimerus sp. genomic DNA was extracted from frozen adult worms that were previously obtained from the livers of naturally infected cats and dogs of Chachi communities, as described elsewhere [4], using a G-spin Total DNA Extraction Kit (Intron Biotechnology) according to the manufacturers’ instructions. DNA was measured using a Nanodrop ND-100 spectrophotometer (Nanodrop Technologies) and then diluted with ultrapure distilled water to final concentrations of 5 ng/μL and 0.5 ng/μL. Serial 10-fold dilutions from adult Amphimerus sp. DNA were prepared with ultrapure water, ranging from 1x10-1 to 1x10-9, and stored at -20°C until use. DNA thus prepared was used as a positive control in all PCR and LAMP reactions as well as for assessing the sensitivity of both molecular assays.
To determine the specificity of PCR and LAMP assays to amplify only Amphimerus sp. DNA, a total of 16 DNA samples from several helminths, including trematodes (Clonorchis sinensis, Opisthorchis viverrini, Fasciola hepatica, Dicrocoelium dendriticum, Schistosoma mansoni, S. haematobium, S. japonicum, and S. intercalatum), cestodes (Echinococcus granulosus and Taenia truncata), nematodes (Onchocerca volvulus, Strongyloides venezuelensis, and Trichinella spiralis) and protozoa (Entamoeba histolytica, Cryptosporidium parvum, and Giardia duodenalis) were used. The concentration of all DNA samples was measured by the same method as described for Amphimerus sp. DNA, which was then diluted with ultrapure water to a final concentration of 0.5 ng/μL and kept at -20°C until use in molecular assays.
Designing LAMP primers
An 459 base pair (bp) sequence, corresponding to a linear genomic DNA partial sequence in the ITS2 region of Amphimerus sp. HS-2011 isolated from human host, was selected and retrieved from GenBank (Accession No. AB678442.1) [4] for the design of the specific primers. The 459 bp sequence was tested using BLASTN analysis [34] for similarity in the available online genome databases. A set of LAMP primers complementary to the nucleotide sequence was designed using the online Primer Explorer V4 software (https://primerexplorer.jp/elamp4.0.0/; Eiken Chemical Co., Ltd., Tokyo, Japan) according to criteria described by Notomi et al [19]. A final complete set of four primers-including a forward outer primer (F3), a reverse outer primer (B3), a forward inner primer (FIP) and a backward inner primer (BIP)-was selected based on the criteria described in “A guide to LAMP primer designing” (http://primerexplorer.jp/e/v4_manual/index.html) of LAMP primers; the locations and target sequence are shown in Fig 1. All the primers were of HPLC grade (Thermo Fisher Scientific Inc., Madrid, Spain). The lyophilized primers were resuspended in ultrapure water to a final concentration of 100 pmol/μL and stored at -20°C until use.
Fig 1. Design of LAMP primers for detecting DNA of Amphimerus sp.
(A) Schematic representation of the 459 bp selected sequence of Amphimerus sp. HS-2011 isolated from human host (AB678442.1). (B) Location of the LAMP primers within the selected sequence. Arrows indicate the direction of extension. (C) Sequences of LAMP primers. F3, forward outer primer; B3, reverse outer primer; FIP, forward inner primer (F1c and F2 sequences); and BIP, reverse inner primer (B1c and B2 sequences).
PCR using outer primers F3 and B3
The outer LAMP primer pair (F3 and B3; Fig 1) was initially tested for Amphimerus sp. specificity by a PCR to verify whether the correct target was amplified. PCR was conducted in 25 μL of a reaction mixture containing 2.5 μL of 10x buffer, 1.5 μL of 25 mmol/L MgCl2, 2.5 μL of 2.5 mmol/L dNTPs, 0.5 μL of 100 pmol/L F3 and B3, 2 U Taq-polymerase and 2 μL (10 ng) of DNA template. Initial denaturation was conducted at 94°C for 1 min, which was followed by a touchdown program for 15 cycles with successive annealing temperature decrements of 1.0°C every 2 cycles. For these 2 cycles, the reaction was denatured at 94°C for 20 s followed by annealing at 64°C-58°C for 20 s and polymerization at 72°C for 30 s. The subsequent 15 cycles of amplification were similar, except that the annealing temperature was 57°C. The final extension was performed at 72°C for 10 min. All PCR reactions were performed in a Mastercycler Gradient-96well (Eppendorf).
The specificity of PCR F3-B3 was tested using heterogeneous DNA samples from other parasites included in the study. The sensitivity was also assayed to establish the detection limit of Amphimerus sp. DNA with 10-fold serial dilutions prepared as mentioned above. All PCR assays were performed with 2 μL of the DNA template (5 ng/μL) in each case. Negative controls (ultrapure water) and positive controls (genomic DNA from Amphimerus sp.) were always included. The PCR products (3–5 μL/each) were subjected to 1.5–2% agarose gel electrophoresis stained with ethidium bromide and visualized under UV light.
Establishing the LAMP assay
We evaluated the LAMP primer set designed by using different reaction mixtures to compare results in Amphimerus sp. DNA amplification. LAMP reactions mixtures (25 μL) contained 40 pmol each of FIP and BIP primers, 5 pmol each of F3 and B3 primers, 1.4 mM each of dNTP (Intron), 1x Isothermal Amplification Buffer-20 mM Tris-HCl (pH 8.8), 50 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.1% Tween20 (New England Biolabs, UK)-betaine (0.8, 1, 1.2, 1.4 or 1.6 M) (Sigma, USA), supplementary MgSO4 (2, 4, 6 or 8 mM) (New England Biolabs, UK) and 8 U of Bst polymerase 2.0 WarmStart (New England Biolabs, UK) with 2 μL (1 ng) of template DNA.
LAMP reactions were performed in 0.5-mL micro-centrifuge tubes that were incubated in a simple heating block at a range of temperatures (61, 63 and 65°C) for 60 min to optimize the reaction conditions and then heated at 80°C for 5–10 min to terminate the reaction. The optimal temperature was determined and used in the following tests. Because of the high sensitivity of the LAMP reaction, DNA contaminations were prevented using sterile tools at all times, performing each step of the analysis in separate work areas and minimizing manipulation of the reaction tubes. Template DNA was replaced by ultrapure water as a negative control in each LAMP reaction.
The specificity of the LAMP assay to amplify only Amphimerus sp. DNA was tested against 16 DNA samples obtained from other parasites used as heterogeneous controls, as mentioned above. To determine the lower detection limit of the LAMP assay, genomic DNA from Amphimerus sp., 10-fold serial diluted as mentioned above, was subjected to amplification compared with the PCR F3-B3.
Detection of LAMP products
The LAMP amplification results could be visually inspected by adding 2 μL of 1:10 diluted 10,000X concentration fluorescent dye SYBR Green I (Invitrogen) to the reaction tubes. Green fluorescence was clearly observed in the successful LAMP reaction, while it remained original orange in the negative reaction. In addition, the LAMP products (3–5 μL) were monitored using 1.5–2% agarose gel electrophoresis stained with ethidium bromide, visualized under UV light and then photographed using an ultraviolet Gel documentation system (UVItec, UK).
Statistical analysis
To estimate the accuracy of the LAMP assay method as a diagnostic test, the percentages of the sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) were calculated using the MedCalc statistical program version 16.8.4 (MedCalc Software, Ostende, Belgium) according to the software instruction manual (www.medcalc.org).
Results
PCR using outer primers F3-B3: Sensitivity and specificity
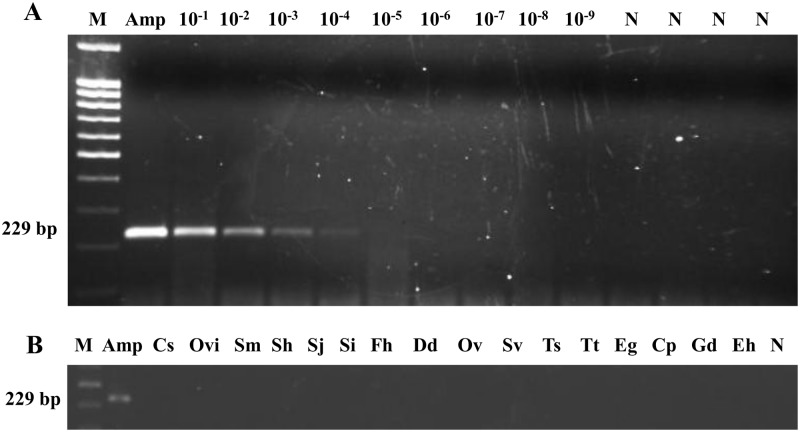
The expected 229 bp PCR product was successfully obtained with outer primers F3 and B3 from Amphimerus sp. DNA. According to sensitivity, the minimum level of Amphimerus sp. DNA detectable by PCR was 0.001 ng (1 pg) (Fig 2A). Additionally, when DNA samples from other parasites included in the study were subjected to this PCR assay, no amplicons were obtained (Fig 2B).
Fig 2. PCR verification, detection limit and specificity using outer primers F3 and B3 for DNA amplification of Amphimerus sp.
(A) Detection limit of PCR F3-B3. Lane Amp, DNA of Amphimerus sp. (10 ng); lanes 10−1–10−9, 10-fold serial dilutions of Amphimerus sp. DNA. (B) Specificity PCR F3-B3. Lanes Amp, Cs, Ovi, Sm, Sh, Sj, Si, Fh, Dd, Ov, Sv, Ts, Tt, Eg, Cp, Gd, Eh: DNA samples of Amphimerus sp., Clonorchis sinensis, Opisthorchis viverrini, Schistosoma mansoni, S. haematobium, S. japonicum, S. intercalatum, Fasciola hepatica, Dicrocoelium dendriticum, Onchocerca volvulus, Strongyloides venezuelensis, Trichinella spiralis, Taenia truncata, Echinococcus granulosus, Cryptosporidium parvum, Giardia duodenalis and Entamoeba histolytica, respectively. In all panels: lane M, molecular weight marker (100 bp Plus Blue DNA Ladder) and lane N, negative controls (ultrapure water, no DNA template).
Examination of human stool samples by PCR F3-B3
We tested the 44 human stool samples by PCR using the outer primers F3 and B3, and very faint bands of the expected size (229 bp) were only obtained in 3 samples (nos. 31, 34 and 45) (S1 Fig).
Establishing the LAMP assay: LAMPhimerus
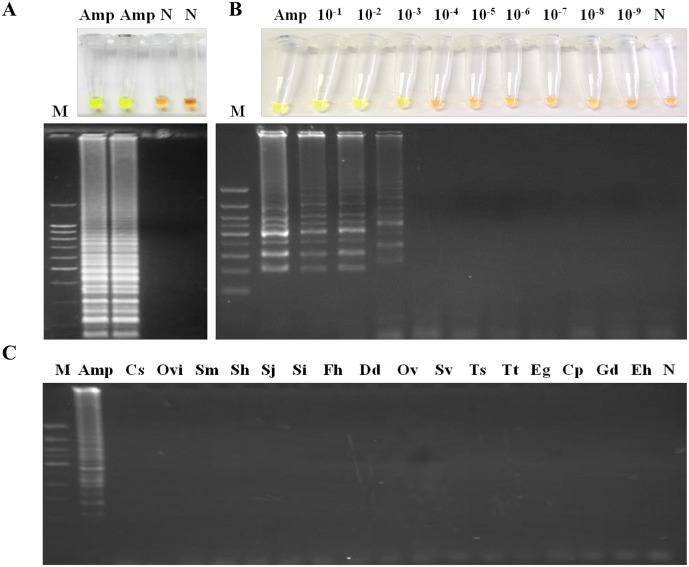
Subsequent to testing different reaction mixtures and temperature conditions, the best amplification results (based on the most evident color change by adding the fluorescent dye and the intensity of the multiple bands on agarose as well as reproducibility of tests) were always obtained when the LAMP master mixture contained 1 M betaine combined with supplementary 6 mM MgSO4 (resulting in a final concentration of 8 mM MgSO4 in 1x Isothermal Amplification Buffer) and was incubated at 63°C for 60 min in a heating block (Fig 3A). When we evaluated the sensitivity of the established LAMP assay, the limit of detection in Amphimerus sp. genomic DNA amplification was identical to that obtained when using PCR with outer primers, specifically 0.001 ng (1 pg) (Fig 3B). To determine the specificity of the LAMP assay for Amphimerus sp., a panel of 16 additional DNA samples from other parasites was tested for amplification. A positive result was only obtained when Amphimerus sp. DNA was used as template, while DNA samples from other specimens were not amplified, demonstrating its high specificity (Fig 3C).
Fig 3. Establishing the LAMP assay.
(A) LAMP amplification results obtained using the established LAMPhimerus assay with the addition of SYBR Green I (up) or visualization on agarose gel (down). Lane M, molecular weight marker (100 bp Plus Blue DNA Ladder); lane Amp, Amphimerus sp. DNA (1 ng); and lane N, negative control (ultrapure water and no DNA template). (B) Sensitivity assessment of LAMPhimerus using serial dilutions of Amphimerus sp. genomic DNA. Lane M, molecular weight marker (100 bp Plus Blue DNA Ladder); lane Amp, Amphimerus sp. DNA (1 ng); lanes 10−1–10−9, 10-fold serial dilutions; and lane N, negative control (ultrapure water and no DNA template). (C) Specificity of the LAMPhimerus assay. Lane M, molecular weight marker (100 bp Plus Blue DNA Ladder); lane Amp, Cs, Ovi, Sm, Sh, Sj, Si, Fh, Dd, Ov, Sv, Ts, Tt, Eg, Cp, Gd, and Eh: DNA samples of Amphimerus spp., Clonorchis sinensis, Opisthorchis viverrini, Schistosoma mansoni, S. haematobium, S. japonicum, S. intercalatum, Fasciola hepatica, Dicrocoelium dendriticum, Onchocerca volvulus, Strongyloides venezuelensis, Trichinella spiralis, Taenia truncata, Echinococcus granulosus, Cryptosporidium parvum, Giardia duodenalis and Entamoeba histolytica, respectively; and lane N, negative control (ultrapure water and no DNA template).
In this way, the best reaction mixture, in addition to the specific primers designed, was established as the most fitting assay for amplification of Amphimerus sp. DNA and was named "LAMPhimerus" in all successive LAMP reactions.
Application of LAMP in human stool samples: LAMPhimerus analysis
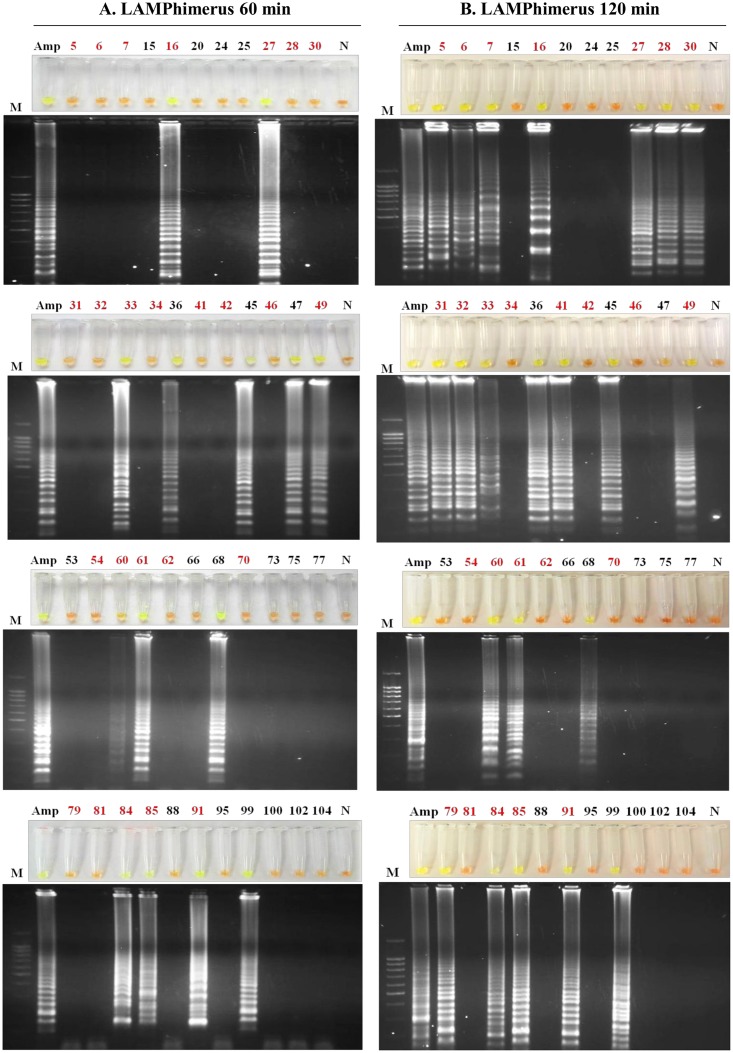
The 44 human stool samples were tested with LAMPhimerus assay using two incubation times for reaction, 60 min and 120 min (Fig 4). To prevent potential cross-contamination, amplification assays were performed in four batches of 11 samples each for easy handling. When testing stool samples using an incubation time of 60 min (Fig 4A), we obtained LAMP positive results in 14/44 (31.81%) samples, including 5 samples (nos. 36, 45, 47, 68 and 99) that were negative in all parasitological tests applied. When using an incubation time of 120 min (Fig 4B), the number of positive results was increased to 22/44 (50%), which also included 4/5 negative parasitological samples as before (nos. 36, 45, 68 and 99). In all LAMP positive reactions, green fluorescence was clearly visualized under natural light conditions. Positive controls always worked well and negative controls were never amplified. All positive results obtained when performing the assay for 60 min were supported at 120 min, except one sample (no. 47). For 120 min, in sample no. 47, a mix between green and orange was observed in the reaction tube; also a very faint smear was visible on agarose gel, indicating poor DNA amplification. Taking together the results obtained from the two incubation assays, we finally considered sample no. 47 as positive, resulting in a total of 23/44 (52.27%) positive LAMPhimerus results.
Fig 4. LAMPhimerus analysis of human stool samples in this study.
(A) Incubation time of 60 min. (B) Incubation time of 120 min. Lane M, molecular weight marker (100 bp Plus Blue DNA Ladder); lane Amp, Amphimerus sp. DNA (1 ng); lane N, negative controls (ultrapure water and no DNA template); and numbers 5–104, analyzed human stool samples. The highlighted red numbers correspond to samples that were positive by one or more applied parasitological methods.
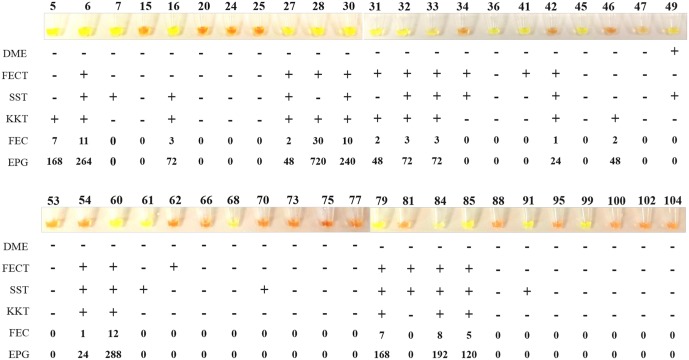
In summary, of the total of 25 parasitologically positive stool samples, we obtained 9/25 (36%) and 18/25 (72%) positive results when we applied LAMPhimerus for 60 min and 120 min, respectively. Additionally, positive results included 5/19 (26.31%) samples (nos. 36, 45, 47, 68 and 99) that were negative in all previously applied parasitological tests. Of the 11 samples (nos. 6, 27, 30, 32, 33, 42, 54, 60, 79, 84, and 85) that were simultaneously positive on three parasitological tests (including the formalin-ether concentration technique, FECT; simple sedimentation technique, SST; and Kato-Katz technique, KKT), 9 (9/11; 82%) were also positive on the LAMP assay; only the 2 samples (nos. 42 and 54) with the same very low egg count (FEC = 1; EPG = 24) were negative on the LAMP assay. Fig 5 shows a comparison of the results obtained for detecting Amphimerus sp. in human stool samples when using the classical parasitological techniques applied and the 120 min-LAMPhimerus assay.
Fig 5. Comparison of the results obtained by the LAMPhimerus assay and classical parasitological techniques applied in this study for detecting Amphimerus sp. in human stool samples.
DME, direct microscopy detection; FECT, formalin-ether concentration technique; SST, simple sedimentation technique; KKT, Kato-Katz technique; FEC, fecal egg count; EPG, eggs per gram of feces; -, negative for egg detection; and +, positive for egg detection. Values indicated for FEC and EPG correspond to the numbers of detected eggs and numbers 5–104 correspond to the analyzed human stool samples.
Considering the results obtained, the following diagnostic parameters for the sensitivity and specificity were calculated for our LAMPhimerus assay: 76.67% sensitivity (95% CI: 52.72% -90.07%); 80.77% specificity (95% CI: 60.65% -93.45%); 82.14% positive predicted value (95% CI: 67.13% -91.20%) and 75.00% negative predicted value (95% CI: 60.43% -85.49%).
Discussion
Human amphimeriasis, caused by the Amphimerus spp. liver fluke, has been recently reported as an emerging zoonotic food-borne trematodiasis [3, 4]. The conventional diagnosis of liver fluke infections in humans is based on the demonstration of eggs in different clinical samples, especially in feces. However, the morphological identification of eggs is troublesome in endemic areas where co-infection with other zoonotic trematodes usually exists. Additionally, stool examination lacks analytical sensitivity, particularly for light infections, requiring serial fecal sampling and an intensive effort in resource-poor settings [35]. To solve these limitations, many immunological and molecular diagnostic approaches have already been developed and applied to detect the presence of several human zoonotic trematode infections with varying accuracy [36, 37].
For detecting Amphimerus spp. infection, conventional coprological techniques are the only ones available, and no immunological or molecular methods have been developed to date. Among the possible molecular methods to be developed, LAMP tests are rapidly becoming an attractive diagnostic option for use under field conditions in laboratories with basic facilities [22, 23]. Hence, in this study, with the aim of improving the diagnostic testing for amphimeriasis, we have developed and evaluated, for the first time, a specific LAMP assay to detect Amphimerus sp. DNA in field samples collected from humans.
At present, nucleotide information for Amphimerus spp. DNA is very scarce, and only a few DNA partial sequences, corresponding to five isolates from hosts (including human, dog, cat, and two softshell turtles), are available in the Genbank database for potential LAMP primers design. The 459 bp sequence of the ribosomal DNA ITS2 region of Amphimerus sp. HS-2011 isolated from human hosts [4] was selected as a target of amplification. This sequence matches those later reported for isolates from a dog (dog-2012) and cat (cat-2012) residing in the same studied endemic indigenous Chachi communities for human amphimeriasis [4]. Therefore, the selection of the target region was appropriate because it seems to contain an identical sequence for all geographical isolates of Amphimerus spp. circulating in the same area, and the assay could be suitable for easily diagnosing both infected animals and humans in endemic areas of amphimeriasis with limited resources.
First, we established the proper operation, sensitivity and specificity of both conventional PCR (using the outer primers) and the LAMP assay (using four specific primers: LAMPhimerus) in the amplification of the Amphimerus sp. DNA target sequence. Both assays were shown to be highly specific for Amphimerus sp. because no cross-reactivity could be observed when DNA from other parasites, including those closely related such as C. sinensis and O. viverrini, were used as a template in the reactions. Identical sensitivities (1 pg of parasite genomic DNA) were obtained for both PCR and LAMPhimerus although the LAMP technique is usually 10–100 fold more sensitive than PCR [38]. However, the sensitivity obtained was the same as that previously reported for O. viverrini detection targeting the ITS1 region in rDNA (ITS1-LAMP) [27, 29] or the mitochondrial nad1 sequence (mito-OvLAMP) [28]. A higher sensitivity (10−5 pg) has been reported for detecting C. sinensis targeting the cathepsin B3 gene [12]. Perhaps, in this study, a greater sensitivity could have been achieved for our LAMPhimerus assay if other DNA target sequences for designing LAMP primers had been available to analyze in databases.
When PCR was specifically tested with the 44 field-collected stool samples, only 3 very faint PCR-positive results were obtained. Varying sensitivity of PCR detection for O. viverrinii [27] and C. sinensis [14] has already been noted when analyzing human stool samples because Taq DNA polymerase inhibitors are frequent in stool specimens. Substances typically present in human feces and dietary components can also limit DNA extraction success [39]. Therefore, improvement of DNA preparation before extraction from stool samples could be a key factor for obtaining better PCR results in Amphimerus sp. DNA detection, as has been previously described for other similar parasites, such as O. viverrini [40, 27]. In our study, the PCR assay is not emphasized because of its very low performance and inconvenience of application in poorly equipped and often short-staffed laboratories in endemic areas.
Better results were obtained when LAMPhimerus method was applied to test human stool samples. A better performance of LAMP assays over conventional PCR methods when analyzing stool samples has been widely reported in the literature because LAMP is more tolerant to sample-derived inhibitors than PCR for diagnostic applications [41, 42]. Therefore, using the initial established reaction time of 60 min, we obtained 14/44 (31.81%) positive results, including 9/25 (36%) that tested positive by microscopy. It has been already suggested that a longer incubation reaction time in the LAMP assay improves the sensitivity and that LAMP negative samples should be incubated longer to reduce false negatives [43]. According to this, a subsequent increase to 120 min of the standard incubation time protocol for the LAMPhimerus assay allowed us to increase the number of positives results up to 23/44 (52.27%), including 18/25 (72%) microscopy-confirmed Amphimerus sp. infections. It should be noted that 5 stool samples with no parasite eggs (nos. 36, 45, 47, 68 and 99) were positive on LAMPhimerus testing regardless of the reaction time used for amplification. These samples could be truly Amphimerus sp. infections that have been microscopically undetected because of the classically low sensitivity of the parasitological diagnosis [35]. Moreover, up to 10 samples without egg counts were also LAMPhimerus positive. This result confirms a greater sensitivity of the LAMPhimerus assay over microscopic examination.
By contrast, 7 truly microscopy Amphimerus-positive samples (nos. 34, 42, 46, 54, 62, 70 and 81) were never amplified. For these samples, values of FEC using the Kato-Katz thick smear method were minimal (between zero and 1–2 eggs), resulting in very low EPG levels. The absence of amplification in these samples was likely not due to the ineffectiveness of LAMPhimerus method because we obtained positive results in other microscopy-positive samples with low EPG levels too. A possibility for the lack of amplification could have been the small quantity (250–300 mg) of the field-collected stool samples finally used for DNA extraction in the laboratory for the LAMP assay. Because eggs of parasites are not equally distributed among the stool specimens [44], it is possible that eggs could have been easily missed in working samples, compromising the Amphimerus sp. DNA obtained and thus subsequent amplification. It is also important to note that we established the minimum amount of Amphimerus sp. genomic DNA detectable by LAMP is 0.001 ng (1 pg). It has been reported that a single egg of a closely related trematode O. viverrini yields 3.72 ng of genomic DNA [45]. Then, theoretically, our LAMP assay would detect Amphimerus sp. DNA corresponding to less than one single egg in a stool sample. Another possibility could have been a mistake in the morphological identification of parasite eggs when performing the stool microscopic examination. This observation would further confirm the specificity of LAMPhimerus method in the amplification of Amphimerus sp. DNA alone.
However, as noted elsewhere, the need for a decision in case management dictates unequivocal result interpretation [22] and some of the drawbacks of LAMP assays, such as potential DNA contamination and carry-over of amplified products when opening the tubes to use the dye, should be considered because they may compromise the test results.
In summary, we have developed, for the first time, a LAMP assay (namely, LAMPhimerus) for the sensitive and specific detection of Amphimerus sp. DNA in human stool samples. After further research for validation, the method could be readily adapted for effective field diagnosis and disease surveillance in amphimeriasis-endemic areas. Future work will be aimed at large-scale studies to further assess the applicability of this novel diagnostic tool.
Supporting information
Analysis of human stool samples included in the study by PCR using outer primers F3 and B3 to detect Amphimerus sp. DNA. In all panels: lane Amp, DNA of Amphimerus sp. (10 ng); lane M, molecular weight marker (100 bp Plus Blue DNA Ladder); lane N, negative control (ultrapure water, no DNA template); and numbers 5–104, stool samples analyzed.
(TIF)
Acknowledgments
The authors would like to thank Dr. Angel Guevara for kindly provided Onchocerca volvulus DNA used in specificity trials.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was funded by the grants from the Universidad Central del Ecuador (CUP 91750000.0000.374072) (WCT) (http://www.uce.edu.ec/) and in part by the Japan Society for the Promotion of Science, JSPS (KAKENHI: Grant No. 25305011) and by grants for Research on Emerging and Re-emerging Infectious Diseases (H23-Shinko-ippan-014 and H26-Shinkoippan-009) from the Ministry of Health, Labor and Welfare of Japan (http://www.mhlw.go.jp/english/). Additional funding was supported by the Health Research Projects: Technological Development Project in Health, grant number DTS16/00207 (AMA) and Health Research Project, grant number PI16/01784 (PFS) of funding institution Instituto de Salud Carlos III (http://www.isciii.es/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Yamaguti S. Synopsis of the digenetic trematodes of vertebrates. Vols. 1 and II Tokyo: Keigaku Co; 1971. p. 1074. [Google Scholar]
- 2.Bowman DD. Amphimerus pseudofelineus (Ward 1901) Barker, 1911 In: Feline clinical parasitology. 1st ed Ames (IA): Iowa State University Press; 2002; p. 151–153. [Google Scholar]
- 3.Calvopiña M, Cevallos W, Kumazawa H, Eisenberg J. High prevalence of human liver infection by Amphimerus spp. Flukes, Ecuador. Emerg Infect Dis. 2011; 17: 2331–2334. doi: 10.3201/eid1712.110373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calvopiña M, Cevallos W, Atherton R, Saunders M, Small A, Kumazawa H, et al. High prevalence of the liver fluke Amphimerus spp. In domestic cats and dogs in an area for human amphimeriasis in Ecuador. PLos Negl Trop Dis. 2015; 9: e0003526 doi: 10.1371/journal.pntd.0003526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization. 2015. “Investing to overcome the global impact of neglected tropical diseases—Third WHO report on neglected tropical diseases”. http://www.who.int/neglected_diseases/9789241564861/en/
- 6.Rim HJ. Clonorchiasis: an update. J. Helminthol. 2005; 79: 269–281. [DOI] [PubMed] [Google Scholar]
- 7.Sripa B, Kaewkes S., Sithithaworn P, Mairiang E, Laha T, Smout M et al. Liver fluke induces colangiocarcinoma. PLos Negl Trop Dis. 2007; 4: 1148–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, EI Ghissassi F, et al. A review of human carcinogens-Part B: biological agents. Lancet. Oncol. 2009; 10: 321–322. [DOI] [PubMed] [Google Scholar]
- 9.Parvathi A, Kumer HA, Prakasha BK, Lu J, Xu X, Hu W, et al. Clonorchis sinensis: development and evaluation of a nested polymerase chain reaction (PCR) assay. Exp Parasitol. 2007; 115: 291–295. doi: 10.1016/j.exppara.2006.09.010 [DOI] [PubMed] [Google Scholar]
- 10.Kim EM, Verweij JJ, Jalili A, van Lieshout L, Choi MH, Bae YM, et al. Detection of Clonorchis sinensis in stool samples using real-time PCR. Ann Trop Med Parasitol. 2009; 103: 513–518. doi: 10.1179/136485909X451834 [DOI] [PubMed] [Google Scholar]
- 11.Rahman SM, Bae YM, Hong ST, Choi MH. Early detection and estimation of infection burden by real-time PCR in rats experimentally infected with Clonorchis sinensis. Parasitol Res. 2011; 109: 297–303. doi: 10.1007/s00436-011-2253-3 [DOI] [PubMed] [Google Scholar]
- 12.Cai XQ, Yu HQ, Bai JS, Tang JD, Hu XC, Chen DH, et al. Development of a TaqMan based real-time PCR assay for detection of Clonorchis sinensis DNA in human stool samples and fishes. Parasitol Int. 2012; 61: 183–186. doi: 10.1016/j.parint.2011.06.010 [DOI] [PubMed] [Google Scholar]
- 13.Sanpool O, Intapan PM, Thanchomnang T, Janwan P, Lulitanond V, Doanh PN, et al. Rapid detection and differentiation of Clonorchis sinensis and Opisthorchis viverrini eggs in human fecal samples using a duplex real-time fluorescence resonance energy transfer PCR and melting curve analysis. Parasitol Res. 2012; 111: 89–96. doi: 10.1007/s00436-011-2804-7 [DOI] [PubMed] [Google Scholar]
- 14.Huang SY, Tang JD, Song HQ, Fu BQ, Xu MJ, Hu XC, et al. A specific PCR assay for the diagnosis of Clonorchis sinensis infection in humans, cats and fishes. Parasitol Int. 2012; 61: 187–190. doi: 10.1016/j.parint.2011.07.010 [DOI] [PubMed] [Google Scholar]
- 15.Wongratanacheewin S, Pumidonming W, Sermswan RW, Pipitgool V, Maleewong W. Detection of Opisthorchis viverrini in human stool specimens by PCR. J Clin Microbiol. 2002; 40: 3879–3880. doi: 10.1128/JCM.40.10.3879-3880.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muller B, Schmidt J, Mehlhorn H. PCR diagnosis of infections with different species of Opisthorchiidae using a rapid clean-up procedure for stool samples and specific primers. Parasitol Res. 2007; 100: 905–909. doi: 10.1007/s00436-006-0321-x [DOI] [PubMed] [Google Scholar]
- 17.Lovis L, Mak TK, Phongluxa K, et al. PCR Diagnosis of Opisthorchis viverrini and Haplorchis taichui Infections in a Lao Community in an Area of Endemicity and Comparison of Diagnostic Methods for Parasitological Field Surveys. J Clin Microbiol. 2009; 47: 1517–1523. doi: 10.1128/JCM.02011-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaewkong W, Intapan PM, Sanpool O, et al. Molecular Differentiation of Opisthorchis viverrini and Clonorchis sinensis Eggs by Multiplex Real-Time PCR with High Resolution Melting Analysis. Korean J Parasitol. 2013; 51: 689–694. doi: 10.3347/kjp.2013.51.6.689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000; 28: E63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, Lowe SB, Gooding JJ. Brief review of monitoring methods for loop-mediated isothermal amplification (LAMP). Biosens Bioelectron. 2014; 61: 491–499. doi: 10.1016/j.bios.2014.05.039 [DOI] [PubMed] [Google Scholar]
- 21.Notomi T, Mori Y, Tomita N, Kanda H. Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. J Microbiol. 2015; 53: 1–5. doi: 10.1007/s12275-015-4656-9 [DOI] [PubMed] [Google Scholar]
- 22.Njiru ZK. Loop-mediated isothermal amplification technology: towards point of care diagnostics. PLoS Negl Trop Dis. 2010; 6: e1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mori Y, Kanda H, Notomi T. Loop-mediated isothermal amplification (LAMP): recent progress in research and development. J Infect Chemother. 2013; 19: 404–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ai L, Li C, Elsheikha HM, Hong SJ, Chen JX, Chen SH, Li X, Cai XQ, Chen MX, Zhu XQ. Rapid identification and differentiation of Fasciola hepatica and Fasciola gigantica by a loop-mediated isothermal amplification (LAMP) assay. Vet Parasitol. 2010; 174: 228–233. doi: 10.1016/j.vetpar.2010.09.005 [DOI] [PubMed] [Google Scholar]
- 25.Cai XQ, Xu MJ, Wang YH, Qiu DY, Liu GX, Lin A, Tang JD, Zhang RL, Zhu XQ. Sensitive and rapid detection of Clonorchis sinensis infection in fish by loop-mediated isothermal amplification (LAMP). Parasitol Res. 2010; 106: 1379–1383. doi: 10.1007/s00436-010-1812-3 [DOI] [PubMed] [Google Scholar]
- 26.Chen Y, Wen T, Lai DH, Wen YZ, Wu ZD, Yang TB, Yu XB, Hide G, Lun ZR. Development and evaluation of loop-mediated isothermal amplification (LAMP) for rapid detection of Clonorchis sinensis from its first intermediate hosts, freshwater snails. Parasitology. 2013; 140: 1377–1383. doi: 10.1017/S0031182013000498 [DOI] [PubMed] [Google Scholar]
- 27.Arimatsu Y, Kaewkes S, Laha T, Hong SJ, Sripa B. Rapid detection of Opisthorchis viverrini copro-DNA using loop-mediated isothermal amplification (LAMP). Parasitol Int. 2012; 61: 178–182. doi: 10.1016/j.parint.2011.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le TH, Nguyen NT, Truong NH, De NV. Development of mitochondrial loop-mediated isothermal amplification for detection of the small liver fluke Opisthorchis viverrini (Opisthorchiidae; Trematoda; Platyhelminthes). J Clin Microbiol. 2012; 50: 1178–1184. doi: 10.1128/JCM.06277-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arimatsu Y, Kaewkes S, Laha T, Sripa B. Specific diagnosis of Opisthorchis viverrini using loop-mediated isothermal amplification (LAMP) targeting parasite microsatellites. Acta Trop. 2015; 141: 368–371. doi: 10.1016/j.actatropica.2014.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen MX, Ai L, Zhang RL, Xia JJ, Wang K, Chen SH, et al. Sensitive and rapid detection of Paragonimus westermani infection in humans and animals by loop-mediated isothermal amplification (LAMP). Parasitol Res. 2011; 108: 1193–1198. doi: 10.1007/s00436-010-2162-x [DOI] [PubMed] [Google Scholar]
- 31.Sierra R. Traditional resource-use systems and tropical deforestation in a multi-ethnic region in North-west Ecuador. Environ Conserv. 1999; 26: 136–145. [Google Scholar]
- 32.Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000; 403: 853–8. doi: 10.1038/35002501 [DOI] [PubMed] [Google Scholar]
- 33.World Health Organization. Basic laboratory methods in medical parasitology. World Health Organization Publications, Geneva, Switzerland, 1992. ISBN 92-4-15410-4. 114 pp. [Google Scholar]
- 34.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990; 215: 403–410. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 35.Johansen MV, Sithithaworn P, Bergquist R, Utzinger J. Towards improved diagnosis of zoonotic trematode infections in Southeast Asia. Adv Parasitol. 2010; 73: 171–195 doi: 10.1016/S0065-308X(10)73007-4 [DOI] [PubMed] [Google Scholar]
- 36.Esteban JG, Muñoz-Antoli C, Toledo R, Ash LR. Diagnosis of human trematode infections. Adv Exp Med Biol. 2014; 766: 293–327. doi: 10.1007/978-1-4939-0915-5_9 [DOI] [PubMed] [Google Scholar]
- 37.Johansen MV, Lier T, Sithithaworn P. Towards improved diagnosis of neglected zoonotic trematodes using a One Health approach. Acta Trop. 2015;141: 161–169. doi: 10.1016/j.actatropica.2013.07.006 [DOI] [PubMed] [Google Scholar]
- 38.Parida M.M, Horioke K, Ishida H, Dash P.K, Saxena P, Jana A.M, Islam M.A, Inoue S, Hosaka N, Morita K. Rapid detection and differentiation of dengue virus serotypes by a real-time reverse transcription-loop-mediated isothermal amplification assay. J. Clin. Microbio. 2005; 43: 2895–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abu Al-Soud W., and Radstrom P.. Effects of amplification facilitators on diagnostic PCR in the presence of blood, feces, and meat. J. Clin. Microbiol. 2000; 38: 4463–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duenngai K, Sithithaworn P, Rudrappa UK, Iddya K, Laha T, Stensvold CR, Strandgaard H, Johansen MV. Improvement of PCR for detection of Opisthorchis viverrini DNA in human stool samples. J Clin Microbiol. 2008; 46: 366–368. doi: 10.1128/JCM.01323-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaneko H, Kawana T, Fukushima E, Suzutani T. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J Biochem Biophys Methods. 2007; 70: 499–501. doi: 10.1016/j.jbbm.2006.08.008 [DOI] [PubMed] [Google Scholar]
- 42.Francois P, Tangomo M, Hibbs J, Bonetti EJ, Boehme CC, Notomi T, et al. Robustness of loop-mediated isothermal amplification reaction for diagnostics applications. FEMS Immunol Med Microbiol. 2011; 62: 41–48. doi: 10.1111/j.1574-695X.2011.00785.x [DOI] [PubMed] [Google Scholar]
- 43.Geojith G, Dhanasekaran S, Chandran S, Kenneth J. Efficacy of loop mediated isothermal amplification (LAMP) assay for the laboratory identification of Mycobacterium tuberculosis isolates in a resource limited setting. J Microbiol Methods. 2011; 84: 71–73. doi: 10.1016/j.mimet.2010.10.015 [DOI] [PubMed] [Google Scholar]
- 44.Levecke B, Behnke JM, Ajjampur SS, Albonico M, Ame SM, Charlier J, et al. A comparison of the sensitivity and fecal egg counts of the McMaster egg counting and Kato-Katz thick smear methods for soil-transmitted helminths. PLoS Negl Trop Dis. 2011; 5: e1201 doi: 10.1371/journal.pntd.0001201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wongsaward Ch, Wongsaward P, Chai J-Y, Paratasilpin T, Anuntalabhochai S. DNA quantities and qualities from various stages of some trematodes using opical and HAT-RAPD methods. Southeast Asian J Trop Med Public Health. 2006; 37: 62–68. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of human stool samples included in the study by PCR using outer primers F3 and B3 to detect Amphimerus sp. DNA. In all panels: lane Amp, DNA of Amphimerus sp. (10 ng); lane M, molecular weight marker (100 bp Plus Blue DNA Ladder); lane N, negative control (ultrapure water, no DNA template); and numbers 5–104, stool samples analyzed.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.