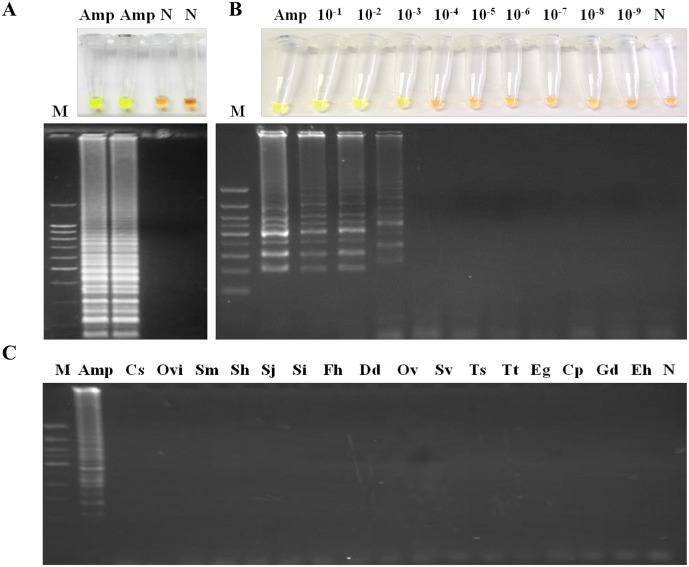
Fig 3. Establishing the LAMP assay.
(A) LAMP amplification results obtained using the established LAMPhimerus assay with the addition of SYBR Green I (up) or visualization on agarose gel (down). Lane M, molecular weight marker (100 bp Plus Blue DNA Ladder); lane Amp, Amphimerus sp. DNA (1 ng); and lane N, negative control (ultrapure water and no DNA template). (B) Sensitivity assessment of LAMPhimerus using serial dilutions of Amphimerus sp. genomic DNA. Lane M, molecular weight marker (100 bp Plus Blue DNA Ladder); lane Amp, Amphimerus sp. DNA (1 ng); lanes 10−1–10−9, 10-fold serial dilutions; and lane N, negative control (ultrapure water and no DNA template). (C) Specificity of the LAMPhimerus assay. Lane M, molecular weight marker (100 bp Plus Blue DNA Ladder); lane Amp, Cs, Ovi, Sm, Sh, Sj, Si, Fh, Dd, Ov, Sv, Ts, Tt, Eg, Cp, Gd, and Eh: DNA samples of Amphimerus spp., Clonorchis sinensis, Opisthorchis viverrini, Schistosoma mansoni, S. haematobium, S. japonicum, S. intercalatum, Fasciola hepatica, Dicrocoelium dendriticum, Onchocerca volvulus, Strongyloides venezuelensis, Trichinella spiralis, Taenia truncata, Echinococcus granulosus, Cryptosporidium parvum, Giardia duodenalis and Entamoeba histolytica, respectively; and lane N, negative control (ultrapure water and no DNA template).