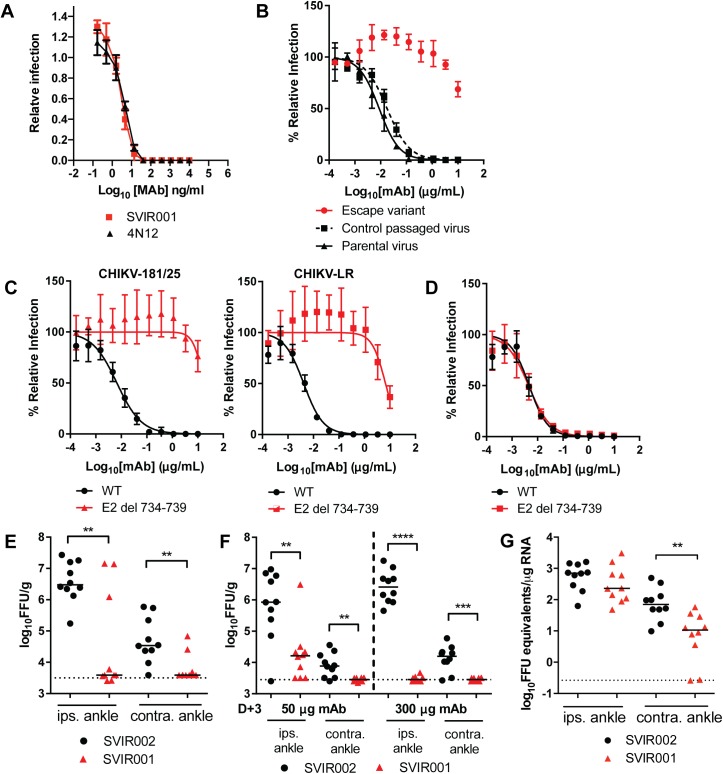
Fig 1. Characterization of neutralization, escape, and therapeutic efficacy of SVIR001 in mice.
(A) SVIR001 and parent 4N12 mAbs were evaluated by neutralization assay in Vero cells. Virus was pre-incubated with indicated concentrations of mAb for 1 h and used to inoculate Vero cells. Data are reported as the relative infection normalized to a no mAb control. Results are representative of one of three independent experiments performed in duplicate. (B) The escape variant virus, which was generated by serial passage in the presence of SVIR001, was subjected to neutralization with SVIR001 and compared to a virus passaged in the absence of SVIR001 (control passaged virus). Results are representative of one of three independent experiments performed in duplicate. (C-D) Confirmation of SVIR001 escape phenotype with engineered six nucleotide deletion (E2 del 734–739). WT or deletion CHIKV-181/25 or CHIKV-LR viruses were incubated with indicated mAb for 1 h. Virus-mAb mixture was added to Vero cells. Data was normalized to a no mAb control. Each graph is two independent experiments done in triplicate and the mean ± SD are shown. (E-G) WT mice were inoculated subcutaneously with 103 FFU of CHIKV-LR in the footpad and treated with 50 μg (E-F) or 300 μg (F-G) of anti-CHIKV mAb SVIR001 or control mAb SVIR002 at 1 (E), 3 (F), or 3 and 10 (G) dpi. At 3 (E), 5 (F), or 28 (G) dpi, virus was quantified by infectious focus (E-F) or qRT-PCR (G) assays from the ipsilateral and contralateral ankle to determine therapeutic efficacy. The median value is shown with the limit of detection indicated by the dotted line. Statistics were calculated on log-transformed data using the Mann-Whitney test (**, P < 0.01, ***, P < 0.001, ****, P < 0.0001). Each data point represents an individual animal. The data (E-G) were pooled from 2 independent experiments.