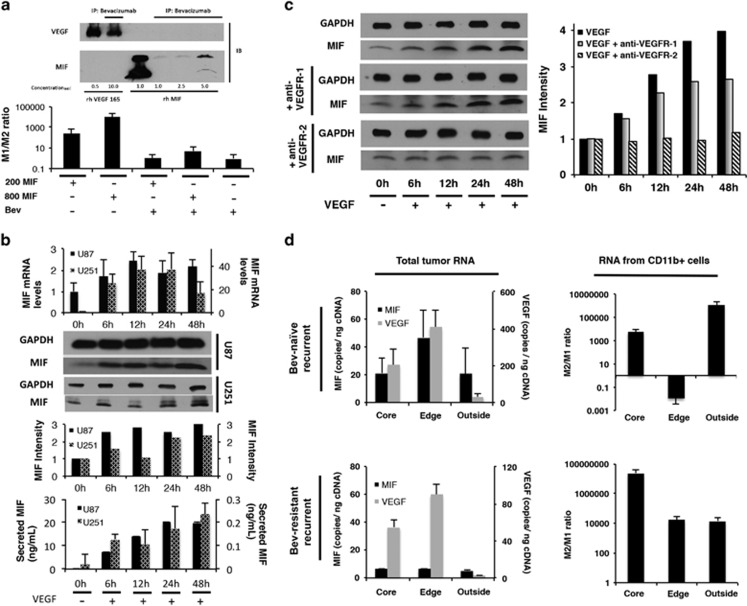
Figure 6.
Dual mechanisms of bevacizumab-induced MIF depletion. (a) Immunoprecipitation revealed that bevacizumab bound MIF, with bevacizumab binding of VEGF serving as a positive control (upper portion of figure). Bevacizumab blocked the ability of recombinant MIF to drive M1 polarization in cultured bone marrow cells (lower portion of figure). (b) Treatment of cultured U87 and U251 cells with 100 ng/ml recombinant VEGF for varying time points up to 48 hours revealed that VEGF increased MIF transcription by qPCR, intracellular protein by western blot of whole cell lysate, and protein secretion by ELISA (P<0.01), offering a potential mechanism by which bevacizumab could reduce tumoral MIF. (c) Cultured U87 cells were treated with VEGF in the presence of blocking antibodies targeting VEGF receptors-1 and 2 (VEGR-1 and VEGFR-2). Blocking VEGFR-2 eliminated VEGF-induced MIF expression, while blocking VEGFR-1 had no effect on VEGF-induced MIF expression. (d) Site-directed biopsies taken from four bevacizumab-naive recurrent glioblastomas revealed increased MIF and VEGF RNA copies at the enhancing edge relative to the central core and FLAIR bright non-enhancing periphery, as assessed by absolute quantification qPCR (P=0.03). In contrast, site-directed biopsies taken from a bevacizumab-resistant recurrent glioblastoma revealed the same regional pattern in VEGF RNA levels, but loss of the spike in MIF RNA that typically occurs at the enhancing edge (P=0.02). Absolute quantification qPCR of column purified CD11b+ cells from bevacizumab-naive versus recurrent glioblastoma revealed elevated M2/M1 ratio at the central core and infiltrated white matter in both tumor types, but at the enhancing edge the bevacizumab-naive tumor had predominantly M1 macrophages, while the resistant tumor had predominantly M2 macrophages (P=0.008-0.01).