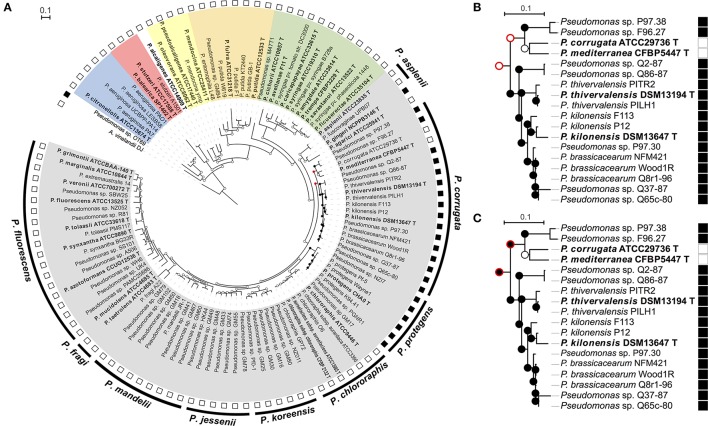
Figure 1.
Ancestral state character reconstruction by Maximum Parsimony of the gains and losses of the phl operon. (A) Phylogenetic rrs-gyrB-rpoB tree of selected Pseudomonas strains in which each node indicates presence (black circle) or absence of phl operon (phlACBD) (white circle). The red circle indicates multiple, equally-parsimonious scenarios that are shown in (B,C). The outer ring represents the subgroups defined in the “Pseudomonas fluorescens” group according to Mulet et al. (2010), the second ring correspond to the presence (black square) or the absence (white square) of the phl cluster. The different Pseudomonas groups are highlighted in color: Gray color, “P. chlororaphis” group; Green, “P. syringae” group; Purple, “P. putida” group; Yellow, “P. oleovorans” group; Red, “P. stutzeri” group; Blue, “P. aeruginosa” group. Fluorescent Pseudomonas strains of uncertain taxonomic status are written as “sp.” and those misclassified were renamed based on rrs-rpoD-gyrB phylogeny and ANI data (See Tables S2–S7). (B,C) The two equally parsimonious scenarios inferred by ancestral reconstruction for the most recent common ancestor of the “P. corrugata” subgroup. Contradicting nodes are denoted by a red outer circle. Strains P97.30, Q37-87, and Q65c-80 likely belong to P. brassicacearum but are portrayed as Pseudomonas sp. in the absence of ANI data.