Figure 2.
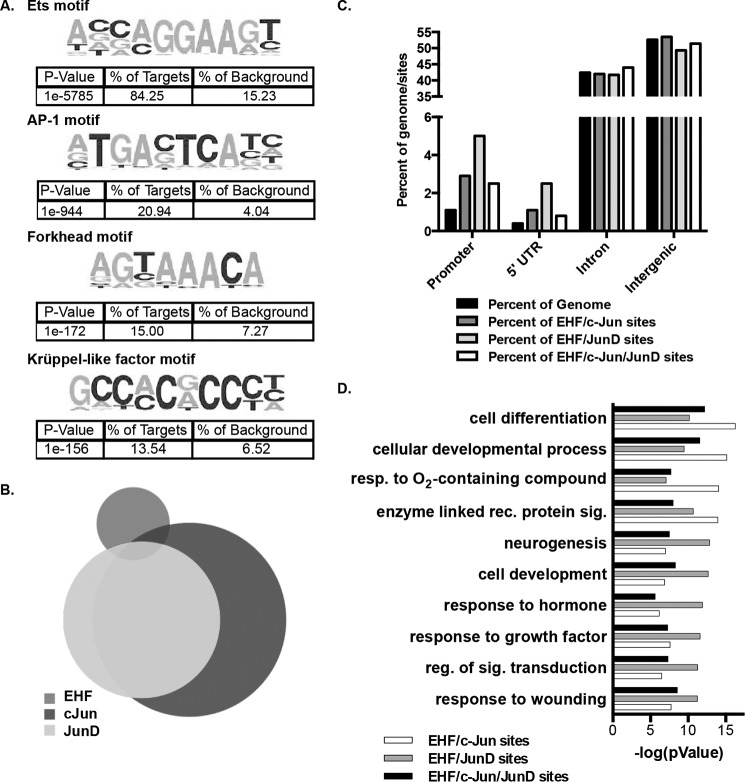
EHF and the AP-1 transcription factor complex bind at overlapping sites genome-wide. A, de novo motif analysis within 200 bp of the center of EHF ChIP-seq peaks in undifferentiated HBE cells identified enriched motifs. The p values, derived by HOMER using cumulative binomial distributions, and the percentage of EHF sites and of background containing each motif are shown below the sequence. B, intersection of EHF, c-Jun, and JunD ChIP-seq binding sites in Calu-3 cells. C, annotation of EHF/c-Jun, EHF/JunD, and EHF/c- Jun/JunD overlapping sites compared with the percentage of the genome. The p values were derived using CEAS (one-sided binomial test). D, gene ontology process enrichment analysis was performed on the nearest gene to each EHF/c-Jun, EHF/JunD, and EHF/c-Jun/JunD intersecting region. Shown are the negative logs of the p values, derived using MetaCore. Resp., response; rec., receptor; sig., signaling; reg., regulation.