Abstract
SERCA1, the sarco(endo)plasmic reticulum Ca2+-ATPase of skeletal muscle, is essential for muscle relaxation and maintenance of low resting Ca2+ levels in the myoplasm. We recently reported that small ankyrin 1 (sAnk1) interacts with the sarco(endo)plasmic reticulum Ca2+-ATPase in skeletal muscle (SERCA1) to inhibit its activity. We also showed that this interaction is mediated at least in part through sAnk1's transmembrane domain in a manner similar to that of sarcolipin (SLN). Earlier studies have shown that SLN and phospholamban, the other well studied small SERCA-regulatory proteins, oligomerize either alone or together. As sAnk1 is coexpressed with SLN in muscle, we sought to determine whether these two proteins interact with one another when coexpressed exogenously in COS7 cells. Coimmunoprecipitation (coIP) and anisotropy-based FRET (AFRET) assays confirmed this interaction. Our results indicated that sAnk1 and SLN can associate in the sarcoplasmic reticulum membrane and after exogenous expression in COS7 cells in vitro but that their association did not require endogenous SERCA2. Significantly, SLN promoted the interaction between sAnk1 and SERCA1 when the three proteins were coexpressed, and both coIP and AFRET experiments suggested the formation of a complex consisting of all three proteins. Ca2+-ATPase assays showed that sAnk1 ablated SLN's inhibition of SERCA1 activity. These results suggest that sAnk1 interacts with SLN both directly and in complex with SERCA1 and reduces SLN's inhibitory effect on SERCA1 activity.
Keywords: calcium ATPase, cytoskeleton, membrane protein, skeletal muscle, transporter, SERCA, sarcolipin, small ankyrin 1
Introduction
The sarco(endo)plasmic reticulum Ca2+-ATPase (SERCA1) facilitates relaxation of skeletal muscle by pumping Ca2+ ions from the cytoplasm into the lumen of the sarcoplasmic reticulum (SR)3 (2, 3). Its function is important to maintain cellular Ca2+ homeostasis and proper excitation-contraction coupling. As several human diseases related to improper Ca2+ handling have been linked to alterations in SERCA expression or activity, the mechanisms regulating SERCA are of great interest (3–7). Much research has been performed to characterize the transmembrane (TM) proteins phospholamban (PLN) and its homologue sarcolipin (SLN) and their ability to inhibit the activity of SERCA (2, 8, 9). Recently, a family of micropeptides related to PLN and SLN has been shown to interact with different SERCA isoforms to regulate their activity in muscle and non-muscle cells (10–12). All of these SERCA regulatory proteins interact with SERCA at least in part through TM interactions that lead to a reduction in SERCA's apparent Ca2+ affinity.
We have recently reported that another small protein, small ankyrin 1 (sAnk1, also known as Ank1.5), can modulate SERCA1. sAnk1 is a 17-kDa TM protein of the nSR in skeletal muscle that, like PLN and SLN, interacts with SERCA1 at least in part via its TM domain to decrease its apparent Ca2+ affinity (1).
Encoded by the ANK1 gene, sAnk1 is member of the ankyrin superfamily and is composed of 155 amino acids (a.a.). It has a unique N terminus (a.a. 1–73) that includes a TM domain (a.a. 1–19) that anchors it within the membrane of the nSR (13–16). The remainder of the protein is exposed in the cytoplasm and includes the last 82 a.a. that share homology to the larger ankyrin isoforms (17, 18). Previous studies have shown that sAnk1 concentrates in the membrane at the level of the M-band and Z-disk, where its C terminus is thought to interact with the myofibrillar proteins obscurin and titin, respectively (19, 20). These interactions are hypothesized to maintain a link between the SR membrane and the underlying proteins of the contractile apparatus. Later studies revealed that elimination of sAnk1 by homologous recombination reduced SR function (21) and that short term reduction of sAnk1 expression by siRNA resulted in disruption of the nSR (17). Reducing sAnk1 expression with siRNA also led to decreased levels of SERCA1 and SLN proteins and inhibition of Ca2+ clearance from the myoplasm (17). These results were among the first to suggest sAnk1 could interact with SERCA1 and SLN.
We recently reported that sAnk1 interacts directly with SERCA1 in a manner similar to, but distinct from, SLN. Similarities between sAnk1 and SLN include their TM a.a. sequences and their shared ability to homodimerize (1, 22, 23). Also, like SLN, sAnk1 binding to SERCA1 leads to a reduction in SERCA1's apparent Ca2+ affinity, mediated by the sAnk1 TM domain.
Intriguingly, the studies of sAnk1 knockdown by Ackermann et al. (17) showed that the residual sAnk1 showed reduced colocalization with SERCA1, whereas its colocalization with SLN was unaffected. Based on this observation, and the ability of SLN and PLN to form homo-oligomers and hetero-oligomers, we asked whether sAnk1 could interact with SLN alone or in a three-way complex with SERCA1. As both sAnk1 and SLN are coexpressed together in skeletal muscle, we hypothesized that the two proteins together may affect SERCA1 activity differently than either protein alone (13, 22), as shown previously for SLN and PLN (24, 25). Here, we investigate whether sAnk1 and SLN interact, and how their coexpression with SERCA1 alters these interactions and the ability to regulate SERCA1 activity.
Results
Interaction of sAnk1 and SLN
coIP was used to study the ability of sAnk1 to interact with SLN in SR vesicles prepared from rabbit skeletal muscle tissue. When antibodies against sAnk1 were used to generate the immunoprecipitate, immunoblot analysis revealed coIP of SLN (Fig. 1A). This suggests that sAnk1 and SLN are present in a complex within the SR membrane.
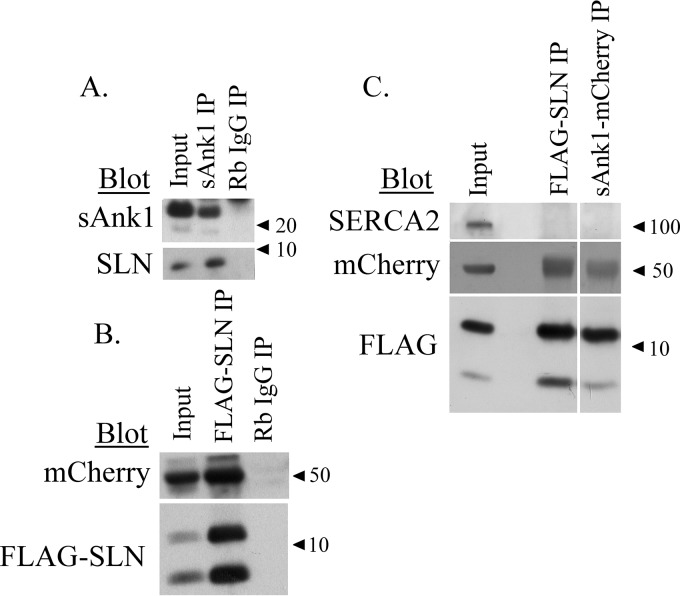
Figure 1.
CoIP of sAnk1 and SLN from rabbit skeletal muscle and COS7 cells. A, SR vesicles were solubilized and subjected to IP with antibodies to sAnk1. After separation by SDS-PAGE, the proteins in the IP were analyzed by immunoblotting with antibodies to sAnk1 and SLN. B, extracts of COS7 cells transfected with sAnk1-mCherry and FLAG-SLN (NF-SLN) at a final plasmid DNA concentration of 1 μg/ml and subjected to IP with antibodies against mCherry. Separated proteins were immunoblotted with antibodies to mCherry and to FLAG. Non-immune rabbit IgG was used as a control in both A and B. The results show that sAnk1 and SLN coIP in skeletal muscle extracts and COS7 cell lysates. C, IP was performed using lysates from COS7 cells transfected as described in B with antibodies against FLAG or mCherry and analyzed by immunoblotting with antibodies against SERCA2, FLAG, and mCherry. The coIP was observed between sAnk1-mCherry and FLAG-SLN, but we did not observe coIP of SERCA2b in either eluate, suggesting that their association does not require SERCA2b.
We next assessed the ability of sAnk1 and SLN to interact in an exogenous expression system. Extracts from COS7 cells transfected to express the sAnk1-mCherry and FLAG-SLN fusion proteins gave similar results. Immunoblots of the IP generated using anti-FLAG showed that sAnk1-mCherry was coeluted with SLN (Fig. 1B), consistent with their interaction in SR vesicles. It is important to note that two bands (∼5 and ∼10 kDa) are observed when immunoblotting for FLAG-SLN. This is likely due to SLN's ability to form stable homodimers that persist even following boiling in solutions containing SDS preparatory to SDS-PAGE (26).
The SERCA2b isoform is expressed in all tissue and cell types (3), including COS7 cells. To learn if sAnk1 and SLN interact directly, and not only in complex with SERCA, we used coIP of the FLAG and mCherry constructs of SLN and sAnk1 to determine whether the endogenous SERCA2b expressed in COS7 cells associated with the sAnk1·SLN complex. As shown in Fig. 1C, SERCA2b fails to coIP with antibodies to either fusion protein. Thus, sAnk1 and SLN associate directly and not via SERCA2b.
Notably, the SLN in the coIPs was primarily dimeric in Fig. 1C but approximately equally distributed between monomers and dimers in Fig. 1B and almost exclusively monomeric in Fig. 1A. The factors that alter the stability of SLN dimers are still poorly understood, but such variability has been reported before (23).
Colocalization of sAnk1-CFP and YFP-SLN in the endoplasmic reticulum (ER) of transfected COS7 cells was demonstrated using confocal microscopy (Fig. 2A). Analysis with Pearson's correlation coefficient showed a similar level of colocalization between sAnk1 and SLN, as we found with SERCA1 and sAnk1 or SLN in our previous study (Fig. 2A; 0.74 ± 0.057) (1). ER localization was confirmed via cotransfection with the ER marker RFP-KDEL, which yielded similar values for Pearson's correlation coefficient (Fig. 2B).
Figure 2.

Colocalization of sAnk1 and SLN and sAnk1, SERCA1, and SLN in COS7 cells. A, COS7 cells were transfected with cDNAs (1 μg/ml final concentration) encoding sAnk1-CFP and YFP-SLN. Significant colocalization was observed, as measured by Pearson's correlation coefficient (included within each merge panel). Values are mean ± S.E. B, additionally, cotransfection of the indicated fluorescent fusion proteins along with an RFP-KDEL marker revealed colocalization of these proteins to the ER. Coefficients in green, blue, and yellow correspond to colocalization between CFP-YFP, CFP-RFP, and YFP-RFP pairs, respectively. Similar results were observed when CFP-SERCA1, sAnk1-YFP, and mCherry-SLN (imaged using identical setting as RFP) were coexpressed. Scale bars, 10 μm.
We previously used AFRET assays to demonstrate the association between sAnk1 and SERCA1 (1). We employed the same methodology here to determine whether Ank1 and SLN reside approximately within 10 nm or less of one another in the ER of COS7 cells. This technique takes advantage of the intrinsically high anisotropy of fluorescent proteins (in contrast to small fluorescent molecules). A donor fluorophore excited with polarized light will have polarized emissions and high anisotropy values. Nearby acceptor molecules may accept photons outside the original plane of polarization via energy transfer, leading to a reduction in the measured anisotropy. This method also reduces the likelihood of false positives caused by overlapping spectra and eliminates the need for other methods of FRET confirmation such as donor dequenching after acceptor photobleaching. For more detail on this method and its advantages, refer to Refs. 27–29.
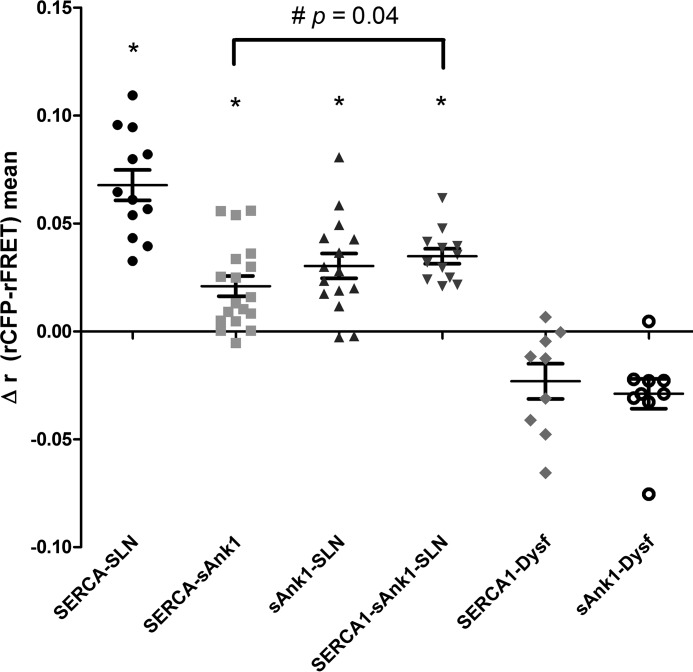
Fig. 3 shows the results of the AFRET experiments. Cells were illuminated with CFP-selective wavelengths, and the fluorescence polarization was measured in both the donor (CFP) and FRET (YFP) channels to look exclusively at heterotransfer FRET between the two fluorescent proteins. YFP anisotropies under direct excitation were also monitored for potential YFP-YFP homotransfer, and anisotropies less than or equal to FRET anisotropies were excluded to minimize the influence of potential homotransfer on the anisotropy measurement in the FRET channel (see under “Experimental procedures”). We found that energy transfer occurred in cells coexpressing sAnk1-CFP and YFP-SLN (Δrmean = 0.03 ± 0.006). This value was statistically significant when tested against a theoretical mean of zero (p = 0.0001). The AFRET value measured for sAnk1-CFP and YFP-SLN was in a range similar to that observed between CFP-SERCA1 and YFP-SLN (Δrmean = 0.068 ± 0.007) and CFP-SERCA1 and sAnk1-YFP (Δrmean = 0.021 ± 0.005). In contrast, dysferlin, a protein of the transverse tubules in skeletal muscle but that accumulates in the ER of COS7 cells, failed to show energy transfer with sAnk1 when it served as the acceptor fluorophore, as reported (1). Calculated anisotropy values are shown in supplemental Fig. 1. The observed FRET between sAnk1 and SLN indicates that these two proteins reside within molecular distances of one another in the ER of COS7 cells, consistent with their direct interaction.
Figure 3.
AFRET of sAnk1 and SLN in COS7 cells. COS7 cells were transfected (1 μg/ml final concentration) with the donor-acceptor pairs (e.g. CFP-SERCA1/YFP-SLN) indicated below the panel. One day post-transfection, AFRET was measured and expressed as Δr (rCFP-rFRET). Each point represents the average AFRET value for a single cell. The t tests for each sample set were performed against a theoretical mean of zero; * indicates that the mean is statistically greater than zero (p < 0.0001). The combinations of sAnk1 and SERCA1 and SLN and SERCA1 were used as positive controls; these data were first reported in Ref. 1. AFRET was observed between sAnk1-CFP and YFP-SLN. When FLAG-SLN was cotransfected with CFP-SERCA1 and sAnk1-YFP, the average AFRET value was significantly increased as measured by t test (#, p = 0.0401).
SLN promotes interaction of sAnk1 and SERCA1
The results presented above raised the question of how sAnk1 and SLN may alter the other's ability to interact with SERCA1. To address this, we first performed coIP experiments with antibodies to SERCA1 in extracts of COS7 cells transfected to express SERCA1 and sAnk1-FLAG or FLAG-SLN, or all three proteins. Intriguingly, the addition of FLAG-SLN led to a dramatic increase in the coIP of sAnk1 with SERCA1 compared with cotransfection of SERCA1 and sAnk1 (Fig. 4A). This increase was statistically significant (2.6-fold; p = 0.0047; Figs. 3 and 4B).
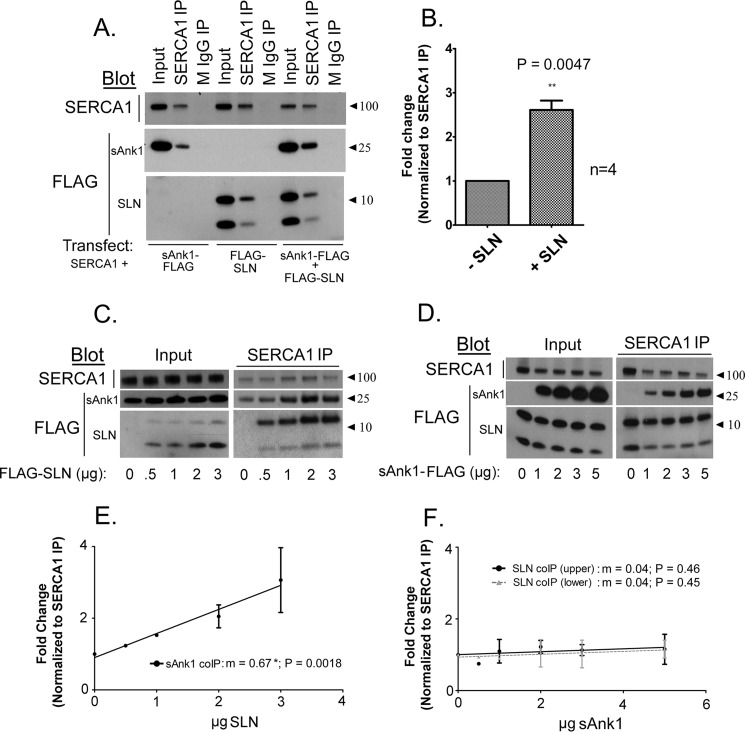
Figure 4.
SLN promotes interaction between SERCA1 and sAnk1. A, COS7 cell extracts transfected as indicated below each panel were subjected to IP with antibodies specific to SERCA1. Transfections were performed using a [C]final of 1 μg/ml. B, quantitative densitometry analysis was performed to assess coIP between SERCA1 and sAnk1 in the presence or absence of FLAG-SLN. There was a 2.6-fold increase in coIP of sAnk1 when it was coexpressed with FLAG-SLN. C, increasing the amount of cDNA encoding FLAG-SLN, as indicated below each panel, increased interaction between SERCA1 and sAnk1. D, increasing the amount of cDNA encoding sAnk1-FLAG had no effect on coIP of SERCA1 and FLAG-SLN. E and F, graphical representation of densitometric analysis of experiments shown in C and D, respectively. Linear regression shows a significant increase in coIP of sAnk1 with SERCA1 with increasing SLN expression (E: m = 0.672 ± 0.15; p = 0.0018; n = 2), whereas increasing sAnk1 expression had no significant effect on coIP of SLN with SERCA1 (F: m = 0.0404 ± 0.53; p = 0.46 (upper band) and m = 0.040 ± 0.05080; p = 0.45 (lower band; n = 2). Data represent slope ± S.E.
Increasing the expression of FLAG-SLN further by introducing greater amounts of FLAG-SLN plasmid during the transfection step promoted increased coIP of sAnk1-FLAG with SERCA1 (Fig. 4, C and E). The increase in sAnk1 present in the coIP varied linearly with the amount of FLAG-SLN cDNA used for transfection and was statistically significant, as determined via linear regression analysis (slope m = 0.67; p = 0.0018). Notably, as seen with coIP of sAnk1 and SLN from extracts of COS7 cells (Fig. 1), SERCA1 also associates preferentially with SLN that appears dimeric after SDS-PAGE, irrespective of the relative amount of associated sAnk1 and despite the fact that most of the SLN in the input lanes appears monomeric.
We next performed the reciprocal experiment, in which we used increasing amounts of sAnk1-FLAG plasmid, while keeping plasmids encoding SERCA1 and FLAG-SLN constant. Unexpectedly, we saw no effect on increasing the amounts of plasmid encoding sAnk1-FLAG on coIP of FLAG-SLN with SERCA1 (Fig. 4D). This was confirmed by linear regression analysis, for which the slope was not significantly different from zero (m = 0.04, both bands; p = 0.46, upper band and 0.45, lower band; Fig. 4F).
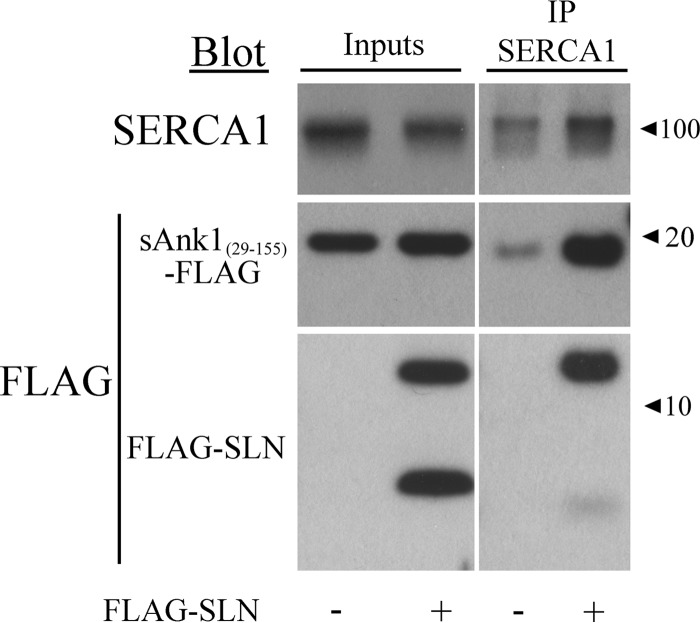
We previously showed that sAnk1 interacts with SERCA1 through both TM and cytoplasmic interactions by using a TM mutant of sAnk1 in which all the residues were changed to leucines and by using a bacterially expressed protein containing the cytoplasmic portion of sAnk1 (sAnk1(29–155)). Here we used the same cytoplasmic region of sAnk1 to determine whether SLN had any effect on the cytoplasmic interaction between sAnk1 and SERCA1. Similarly to its effect on the interaction between SERCA1 and full-length sAnk1, the presence of FLAG-SLN enhanced the interaction between sAnk1(29–155) and SERCA1 by 2.7-fold (Fig. 5).
Figure 5.
SLN promotes interaction between sAnk1's cytoplasmic domain (sAnk(29–155)) and SERCA1. Extracts from COS7 cells were transfected to express SERCA1 and sAnk1(29–155)-FLAG in the presence or absence of FLAG-SLN (NF-SLN). Empty FLAG vector was cotransfected for samples without FLAG-SLN. Transfections were performed using a [C]final of 1 μg/ml. Cell extracts were subjected to IP with antibodies against SERCA1 and immunoblotted. Non-immune mouse IgG was used as a control (data not shown). The results show that the interaction between SERCA1 and the cytoplasmic domain of sAnk1 is increased in the presence of SLN.
We also used AFRET to determine whether FLAG-SLN enhanced the interaction between sAnk1 and SERCA1. Compared with the AFRET signal from COS7 cells transfected with cDNAs encoding CFP-SERCA1 and sAnk1-YFP, the signal from cells cotransfected to express these two proteins and FLAG-SLN had an increased Δrmean (0.021 ± 0.005 versus 0.036 ± 0.003; p = 0.04; Fig. 3). Thus, all our results indicate that SLN promotes the interaction between sAnk1 and SERCA1.
SLN, sAnk1, and SERCA1 interact to form a three-way complex
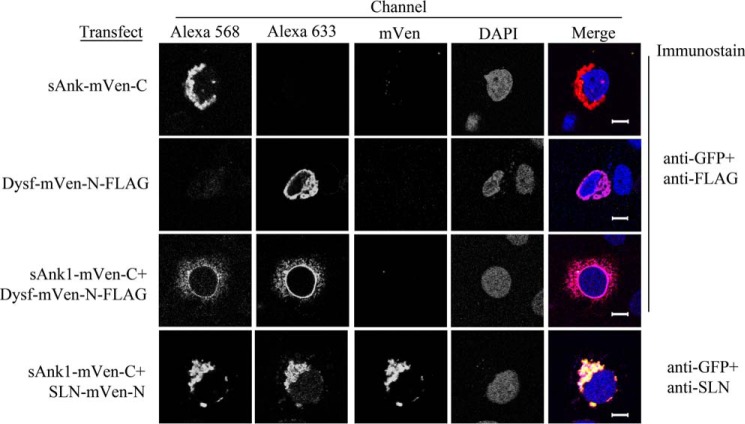
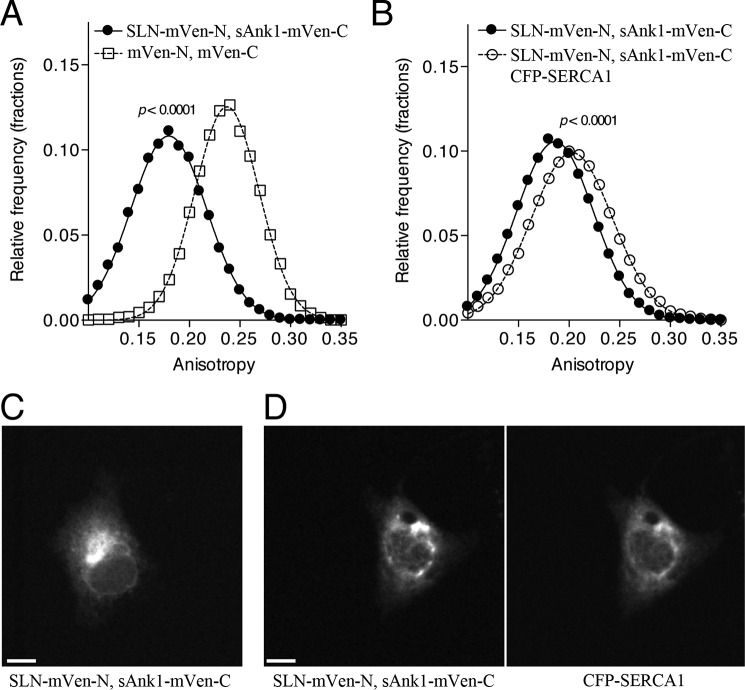
To examine the possibility of a trimeric complex of SLN, sAnk1, and SERCA1, we used a bimolecular fluorescence complementation strategy (30). N- and C-terminal portions of mVen were fused to SLN and sAnk1, respectively, and expressed in COS7 cells. The two split mVen constructs associated to generate a complex that was fluorescent, consistent with the formation of a complete Venus molecule (Fig. 6, 4th row). This was specific, as a small portion of dysferlin, FLAG-tagged and linked to mVen-N, did not generate Venus fluorescence when cotransfected with sAnk1-mVen-C (Fig. 6, 3rd row). Untransfected cells and cells transfected with only one plasmid were used as controls and also showed no Venus fluorescence (Fig. 6, 2nd and 3rd rows and data not shown) This confirms the specific association of sAnk1 with SLN in COS7 cells. Unexpectedly, fluorescence from SLN-mVen-N·sAnk1-mVen-C complexes was highly depolarized (Fig. 7A), indicating a high degree of FRET between SLN·sAnk1 complexes from multimerization. Fluorescence anisotropies of untagged mVenus fragments were consistent with other monomeric fluorescent proteins (31).
Figure 6.
Fluorescence complementation assay in COS7 cells. COS7 cells were transfected with cDNAs encoding sAnk1-mVen-C, SLN-mVen-N, and Dysf-mVen-N-FLAG (1 μg/ml total) as indicated on the left and immunostained as indicated on the right. Alexa 568 secondary antibody was used to detect anti-GFP, and Alexa 633 was used to detect anti-FLAG. All samples were counterstained with DAPI to visualize nuclei. Individual expression of sAnk1-mVen-C (1st row), Dysf-mVen (2nd row), or both together (3rd row) demonstrate that these constructs are expressed in the ER of the COS7 cells but do not give a Venus signal. Cells coexpressing sAnk1-mVen-C and SLN-mVen-N (4th row) demonstrate Venus fluorescence. Scale bars, 10 μm.
Figure 7.
SERCA1 affects SLN-sAnk1 interactions. A, COS7 cells were cotransfected with either SLN-mVen-N and sAnk1-mVen-C or control plasmids containing only the two half-fragments of mVenus (mVen-N and mVen-C). Transfections were performed using a [C]final of 1 μg/ml. Cells were imaged using fluorescence polarization microscopy, and anisotropy values were tabulated at the pixel level. Histograms were normalized to the total number of pixels (mVen-N + mVen-C, 13,322 pixels, ∼40 cells; SLN-mVen-N + sAnk1-mVen-C, 22,790 pixels, ∼150 cells; p < 0.0001 by t test; curves shown are fits to a single component Gaussian (r2 = 0.9990 for each curve)). B, cotransfection with CFP-SERCA1 changed the distribution of anisotropy values observed for SLN-mVen-N·sAnk1-mVen-C FRET (SLN-mVen-N + sAnk1-mVen-C, 36,623 pixels, ∼270 cells; SLN-mVen-N + sAnk1-mVen-C + CFP-SERCA1, 11,898 pixels, ∼100 cells, p < 0.0001 by t test; curves shown are fits to a single Gaussian component (r2 = 0.9997 and 0.9993 for −/+ CFP-SERCA1, respectively)). C and D show representative images for SLN-mVen-N + sAnk1-mVen-C in the absence (C), and presence (D) of CFP-SERCA1. Scale bars, 10 μm.
Next, we tested whether coexpression of CFP-SERCA1 affected the interaction of the SLN·sAnk1 multimeric complex (Fig. 7B). Expression of CFP-SERCA1 shifted the distribution of anisotropy values observed for SLN-mVen-N·sAnk1-mVen-C complexes collected under YFP illumination to examine homotransfer FRET. Notably, the anisotropy values under this condition are depolarized compared with mVen-N and mVen-C complexes alone, indicating the presence of FRET, albeit significantly reduced compared with conditions lacking CFP-SERCA1. This result indicates that the SLN-mVen-N·sAnk1-mVen-C complexes remain intact, but their relative molecular orientation becomes shifted after association with CFP-SERCA1. This is consistent with the formation of a three-way complex containing sAnk1, SLN, and SERCA1. Notably, the anisotropy did not increase to monomeric levels observed in the untagged mVen-N, mVen-C control, suggesting that the complexes are still intact and within molecular distances. We also did not note a change in the expression pattern of SLN-mVen-N·sAnk1-mVen-C in the presence of SERCA1 (Fig. 7, C and D), and all constructs localized strongly to the perinuclear ER region.
sAnk1 reduces SLN-mediated SERCA1 inhibition
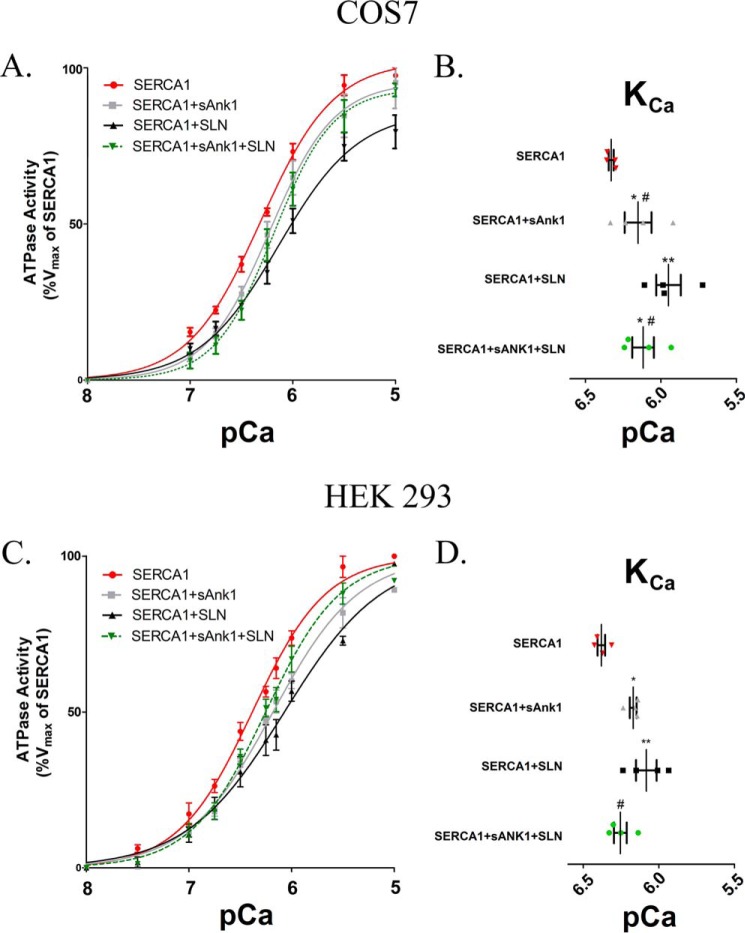
We next studied the effects on SERCA1's Ca2+-ATPase activity in the presence of sAnk1 and SLN expressed together, compared with its activity with either protein alone. Our assays used microsomes prepared from COS7 or HEK293 cells and a colorimetric method to measure Pi release. Cells transfected with SERCA1 alone served as a control and showed the highest level of Ca2+-dependent ATPase activity in both cell types (COS7, pCa = 6.33 ± 0.016; HEK293, pCa = 6.38 ± 0.025). As reported previously (1), sAnk1-FLAG reduced SERCA1's apparent Ca2+ affinity, measured in pCa units, in COS7 and HEK293 cells (ΔKCa2+ = −0.18 and −0.21, pCa2+ units, respectively; Fig. 8 and Tables 1 and 2). This shift was less than that produced by coexpression of FLAG-SLN (ΔKCa2+ = −0.38 pCa2+ units in COS7 and −0.30 in HEK293; Fig. 8; Tables 1 and 2). When sAnk1-FLAG and FLAG-SLN were coexpressed with SERCA1, the level of inhibition was less than that exhibited by FLAG-SLN alone (ΔKCa2+ = −0.21 in COS7 and −0.12 pCa2+ units in HEK293; Fig. 8; Tables 1 and 2) and was statistically identical to that seen with sAnk1 and SERCA1. These findings suggest that sAnk1 reduces SLN-mediated SERCA1 inhibition and may ablate it completely.
Figure 8.
Ca2+-ATPase assays. COS7 (A) and HEK293 (C) cells were transfected with the indicated cDNA construct(s) using a [C]final of 1 μg/ml. ATPase activity was determined at each [Ca2+]free compared with the Vmax measured for SERCA1 alone, following normalization of the levels of SERCA1 expression as determined by immunoblotting (see “Experimental procedures”). Data were fitted to the equation for a general cooperative model for substrate binding. Results from both cell lines show that coexpression of sAnk1 with SERCA1 leads to a reduction of SERCA1's apparent affinity for Ca2+, but the effect of sAnk1 is less than that of SLN, as shown previously (1). B and D, KCa ([Ca2+]free required for half-maximal activation) values were determined from each curve and are summarized in Tables 1 and 2. COS7 mean KCa: SERCA1 pCa = 6.33 (468 nm); SERCA1 + sAnk1 pCa = 6.15 (708 nm); SERCA1 + SLN pCa = 5.95 (1122 nm); and SERCA1 + sAnk1 + SLN pCa = 6.12 (759 nm). HEK293 mean KCa: SERCA1 pCa = 6.38 (415 nm); SERCA1 + sAnk1 pCa = 6.17 (680 nm); SERCA1 + SLN pCa = 6.08 (830 nm); and SERCA1 + sAnk1 + SLN pCa = 6.26 (560 nm). Statistics used one-way ANOVA: *, p < .05 versus SERCA1; **, p < .01 versus SERCA1; and #, p < .05 versus SERCA1 + SLN. † indicates data presented previously (1) and performed at same time as SERCA1 + sAnk1 + SLN.
Table 1.
ATPase assays
COS7 ATPase assays.
The results from ATPase assays presented in Fig. 6 for COS7 cells are summarized. The KCa2+ ([Ca2+] resulting in half-maximal activation) is given in pCa units (right-hand column) and nanomolar concentration (left-hand column). The change in KCa2+ (ΔKCa2+) relative to control (SERCA1 alone) is given in pCa units. Results are mean values ± S.E. NA means not applicable.
|
KCa2+ |
KCa2+ | n | ||
|---|---|---|---|---|
| nm [C] | pCa2+ | |||
| SERCA1 alone | 468 ± 17 | 6.33 ± .016a | NA | 4 |
| SERCA1 + sAnk1 | 708 ± 130 | 6.15 ± .089a,b,c | −0.18 | 4 |
| SERCA1 + SLN | 1122 ± 190 | 5.95 ± .081a,b | −0.38 | 4 |
| SERCA1 + sAnk1 + SLN | 759 ± 120 | 6.12 ± .071b,c | −0.21 | 4 |
a Data were previously shown in Ref. 1. All experiments presented were performed at same time.
b Data are significantly different from SERCA1 alone.
c Data are significantly different from SERCA1 + SLN.
Table 2.
ATPase assays
HEK293 ATPase assays.
The results from ATPase assays presented in Fig. 6 for HEK293 cells are summarized. given in pCa units (right-hand column) and nanomolar concentration (left-hand column). The change in KCa2+ (ΔKCa2+) relative to control (SERCA1 alone) is given in pCa units. Results are mean values ± S.E. NA means not applicable.
|
KCa2+ |
KCa2+ | n | ||
|---|---|---|---|---|
| nm [C] | pCa2+ | |||
| SERCA1 alone | 415 ± 23 | 6.38 ± .025 | NA | 4 |
| SERCA1 + sAnk1 | 680 ± 34 | 6.17 ± .022a | −0.21 | 4 |
| SERCA1 + SLN | 830 ± 127 | 6.08 ± .068a | −0.3 | 4 |
| SERCA1 + sAnk1 + SLN | 560 ± 52 | 6.26 ± .042b,c | −0.12 | 4 |
a Data are significantly different from SERCA1 alone.
b Data are significantly different from SERCA1 + SLN.
c Data are not significantly different from SERCA1 alone.
Discussion
sAnk1 and SLN share several features that suggest that they could interact in the membrane of the nSR and regulate SERCA1 activity in unique ways. First, their TM sequences are highly similar (1), and both proteins homo-oligomerize, SLN primarily through interactions of its TM domain (22, 23). SLN also hetero-oligomerizes with its homologue PLN, which also shares sequence similarity with sAnk1 (1, 9). Furthermore, partial knockdown of sAnk1 by siRNA shows that the remaining sAnk1 colocalizes with SLN in small membrane compartments, presumed to be fragments of the nSR. This colocalization was greater than that observed between sAnk1 and SERCA1 (17). Like SLN and PLN, sAnk1 interacts with SERCA1 to reduce its affinity for Ca2+ (2, 9) and requires its TM domain for this inhibitory activity. Finally, SLN and PLN can combine to super-inhibit SERCA1 activity, presumably by forming a complex containing all three proteins (25, 32). These observations suggested that sAnk1 would interact with SLN and that this interaction would alter SERCA1 activity in a distinct way. Here, we show that sAnk1 can indeed interact with SLN. When coexpressed with SERCA1, SLN promotes interaction between sAnk1 and SERCA1. Surprisingly, unlike PLN, which is super-inhibitory when expressed together with SLN, sAnk1 suppresses SLN's ability to inhibit the Ca2+-ATPase activity of SERCA1. As sAnk1 and SLN are coexpressed and colocalize with SERCA1 in the nSR, our results suggest that their interactions regulate Ca2+ homeostasis in skeletal muscle.
We first investigated whether sAnk1 and SLN interact with one another by using coIP and AFRET experiments, as we did to characterize the interaction between SERCA1 and sAnk1 (1). The coIP studies revealed that sAnk1 and SLN were able to associate specifically in SR vesicles isolated from rabbit back and hind limb muscles and in extracts of transfected COS7 cells. We also showed that sAnk1-CFP and YFP-SLN fusion proteins exhibited energy transfer in AFRET in the ER membranes of COS7 cells, consistent with their close interaction.
Together, these data suggest that SLN and sAnk1 can associate, but they do not prove that their association is direct and not mediated by SERCA2b. Mediation by endogenous SERCA2b is unlikely, however, as we found that SERCA2b does not coIP with either FLAG-SLN or sAnk1-mCherry when they were coexpressed in COS7 cells (Fig. 1C). This is consistent with the observations of Asahi et al. (25), who found no appreciable coIP of SERCA2b with either PLN or SLN, although the two small proteins associated with each other avidly.
We next questioned how the interaction of sAnk1 and SLN would affect either the protein's association with SERCA1 or the regulation of SERCA1's Ca2+-ATPase activity. We addressed the question of association using both coIP and AFRET with COS7 cells transfected to express SERCA1, sAnk1-FLAG, and FLAG-SLN. Compared with cells expressing only SERCA1 and sAnk1, those containing all three proteins showed a significant increase in the level of coIP and energy transfer between sAnk1 and SERCA1. When coIP experiments were performed using increasing amounts of FLAG-SLN cDNA, sAnk1 in the pellet generated with antibodies to SERCA1 increased linearly with the expression of FLAG-SLN, suggesting that SLN promotes the association of sAnk1 and SERCA1. Furthermore, we found that SLN was also able to promote the interaction between SERCA1 and the cytoplasmic portion of sAnk1 (sAnk1(29–155)). By contrast, the coIP of SLN with SERCA1 did not increase with increasing expression of sAnk1, suggesting that the effects of these two proteins on their association with SERCA1 are distinct. Together, these results suggest that SLN can promote the specific interaction of sAnk1 with SERCA1 but that sAnk1 fails to promote SLN-SERCA1 interactions. This result is consistent with those of Asahi and co-workers in a series of publications in 2002 and 2003 (25, 33, 34), which showed that coIP of SERCA1 with PLN increased 3-fold when the two proteins were coexpressed with SLN but that coIP of SLN with SERCA1 was not changed by PLN. These results are strikingly similar to those we observe with SERCA1 and sAnk1.
Our results strongly suggest that SLN, sAnk1, and SERCA1 form a three-way complex. We tested this by examining AFRET between CFP-SERCA1 and Venus moieties formed by SLN linked at its N-terminal fusion protein with the N-terminal portion of Venus and sAnk1 linked at its C terminus to the C-terminal region of Venus. We showed that, when expressed together, the split Venus constructs generated Venus fluorescence and that this was specific, as replacement of the SLN construct with a small transmembrane portion of dysferlin did not produce Venus fluorescence. Remarkably, SLN·sAnk1-Venus displayed a large amount of homotransfer to other SLN·sAnk1-Venus protein complexes. The anisotropy was also strongly depolarized compared with mVenus formed from mVen-N and mVen-C control proteins. As FRET is the only possible mechanism for fluorescence depolarization of large fluorophore fluorescence, these results indicate the formation of a multimeric complex containing multiple SLN·sAnk1 molecules. Interestingly, addition of SERCA1 shifted the anisotropy of the SLN·sAnk1 fluorescence. As these cells retain Venus fluorescence, SERCA1 binding did not dissociate the SLN·sAnk1 complex. Furthermore, the anisotropy levels observed are far closer to the non-SERCA1 conditions than to the control Venus anisotropy, indicating that a large amount of SLN·sAnk1 Venus FRET remains. The simplest interpretation of these findings is that SERCA1 directly interacts with SLN·sAnk multimers in a way that alters their physical positions within a three-way complex, but it does not destabilize the multimers.
It remains unclear how SLN promotes the interaction between SERCA1 and sAnk1. One possible explanation is that SLN induces a conformational change in one or more of the structural domains of SERCA1. If the conformation of the SLN-bound Ca2+ pump opens a new site for sAnk1 to bind, this could explain the observed increase in coIP and energy transfer. It would also explain the increased association of the cytoplasmic domain of sAnk1 with SERCA1 that we observed in the presence of SLN. This possibility seems unlikely, however, as any effect of SLN on the apparent affinity of sAnk1 for SERCA1 should be reciprocal. If this is the mechanism by which SLN promotes sAnk1-SERCA1 binding, it would likely not be explained by traditional allosteric theory.
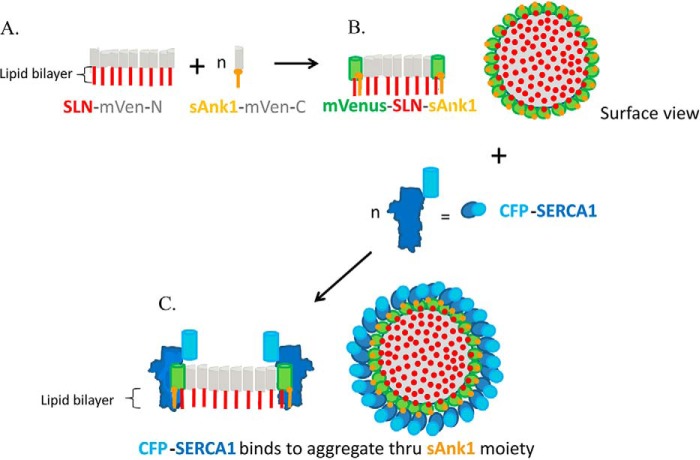
Alternatively, oligomers of SLN could promote SERCA1-sAnk1 interaction. In this scenario, large oligomers of SLN would form preferentially in the ER or SR membrane, as suggested by earlier reports (23, 26). As SLN associates with itself more avidly than it associates with SERCA1 (23), SLN homo-oligomers would be favored over SLN-SERCA1 heterodimers, especially at high relative levels of SLN. SLN oligomers would have a higher apparent affinity for sAnk1 and SERCA1 than SLN monomers, perhaps accumulating them at the exposed rim of the oligomer. Thus, both proteins would selectively associate with SLN oligomers, either alone or together. SERCA1 and sAnk1 (or PLN, in the case of Asahi et al. (25, 33, 34)) would remain monomeric or dimeric (sAnk1 forms stable homodimers (22), and so would concentrate at the rim of the oligomer, perhaps without promoting further SLN oligomerization (Fig. 9). This model can account for our observations with coIP by anti-SERCA1 of sAnk1 and SLN as a function of increasing expression of each, and would be consistent with an approximately linear dependence of the coIP of sAnk1 with SERCA1 as a function of SLN levels, as observed in Fig. 4E (see supplemental Fig. 2 and legend). It would also account for the ability of sAnk1 to ablate SLN's inhibition of SERCA1 as well as for the observations of Asahi and co-workers (25) with PLN and SLN. Finally, it would be consistent with our observation that both SERCA1 and sAnk1 prefer to associate with SLN dimers over monomers in coIP studies (Figs. 4 and 5). Autry et al. (23) reported the formation of oligomers of SLN but argued that the monomer, not dimers or oligomers, associated with SERCA as measured by FRET. We are currently exploring experimental conditions designed to reconcile our results with those of Autry et al. (23).
Figure 9.
Model of complex formed by SLN oligomers, sAnk1 and SERCA1. The model assumes that SLN oligomerizes with itself much more avidly than it hetero-oligomerizes with either sAnk1 or SERCA1. It also assumes that sAnk1 and SERCA1 do not form large aggregates on their own or together, but instead they interact with SLN molecules at the edges of the SLN aggregates. Thus, when SLN is expressed as N-terminal half-Venus chimeras in COS7 cells, it forms large aggregates in the ER (shown in A). sAnk1, expressed as C-terminal half-Venus chimeras, heterodimerizes with SLN-half-Venus moieties at the edge of the SLN aggregates (shown in B in cross-section and surface view), resulting in the reconstitution of Venus and the appearance of Venus fluorescence (shown in green). Expression of SERCA1-Cerulean (shown in blue) together with sAnk1 and SLN leads to its association through sAnk1 with the SLN aggregates (C). The model accounts for the three-way complexes formed by sAnk1, SLN, and SERCA1, the ability of sAnk1 to ablate the inhibitory activity of SLN on SERCA1 activity, the high anisotropy of the SLN-sAnk1 complexes, and the small shift that occurs in the presence of SERCA1. It also accounts for the observation that the amount of sAnk1 increases linearly with SLN expression in the SERCA1 coIP, but not vice versa, as the number of sAnk1-half-Venus molecules at the perimeter of the SLN aggregate varies approximately linearly with the number of half Venus-SLN molecules when the latter is limited (25 < n < 350; see supplemental Fig. 2). Figure was not drawn to scale.
Although the exact dynamics of the interactions among sAnk1, SLN, and SERCA1 remain uncertain, it is clear that the three proteins coordinately regulate Ca2+-ATPase activity. ATPase assays revealed that sAnk1 ablates SLN-mediated SERCA1 inhibition. The presence of sAnk1 ensures that SLN cannot further reduce SERCA1's apparent Ca2+ affinity. The effect of all three proteins on SERCA1 activity was statistically indistinguishable from the effect of sAnk1 alone. Future experiments to determine the specific moieties that mediate the interactions of these three proteins will be important to elucidate the mechanism behind sAnk1's ability to reduce SLN-mediated SERCA1 inhibition and to dissect the role of the three-way complex in vivo. Furthermore, as the levels of SLN and SERCA1 are altered in the mouse model of Duchenne muscular dystrophy (35), it will be of considerable interest to learn whether the levels of sAnk1 are modified in ways that might contribute to pathogenesis.
Experimental procedures
Materials
The chemiluminescence kit used for immunoblotting was from Applied Biosystems (Foster City, CA). ATP, A23187, and thapsigargin were from Sigma. Lipofectamine and Dynabeads coupled with sheep anti-rabbit IgG or sheep anti-mouse IgG were from Invitrogen. The Pi ColorLock ALS reagent was from Novus Biologicals (Littleton, CO). Buffers were supplemented with Complete Protease Inhibitor Mixture from Roche Applied Science.
Antibodies
Primary antibodies against SLN were made by injecting rabbits with the C-terminal sequence of SLN (acetyl-LVRSYQYC-amide and C-aminohexanoic acid-LVRSYQY-OH) linked to bovine serum albumin (BSA). Antibody generation and affinity purification were carried out by 21st Century Biochemicals, Inc. (Marlborough, MA). Primary antibodies against sAnk1 were generated as described previously (1). Other antibodies used include the following: SERCA1 (IIH11 mAb) and SERCA2 (2A7-A1) from Thermo Fisher Scientific; FLAG (M2 mAb and rabbit pAb, Sigma); mouse IgG1κ (MOPC-21, Sigma); rabbit IgG (Jackson ImmunoResearch (West Grove, PA)); and SLN (rabbit pAb, Proteintech Group (Chicago)).
cDNAs
Constructs encoding SERCA1-pcDNA3.1, CFP-SERCA1, YFP-SERCA1, YFP-SLN, FLAG-SLN, sAnk1-FLAG, CFP-sAnk1, YFP-sAnk1, and YFP-dysferlin were generated as described previously (1). Construction of sAnk1-mCherry was previously described (17). The DS-Red-KDEL construct was a gift from Dr. S. Fang, Dept. of Physiology, University of Maryland, Baltimore. We use the generic terms CFP, YFP, and RFP henceforth when referring to constructs made using the pmCerulean3-C1, pmVenus-C1, and DS-Red-KDEL vectors, respectively.
The mVen vectors used for fluorescence complementation experiments were constructed using the mVen-C-N1 and mVen-N-C1 vectors that code for the C- and N-terminal portions of mVen, respectively (gifts from M. Frohman, Stony Brook University). sAnk1-mVen-C was generated by inserting the sAnk1 open reading frame into the mVen-C-N1 vector between the EcoRI (5′) and KpnI (3′) restriction sites. SLN-mVen-N was generated using oligomers spanning the entire rabbit SLN-coding region flanked with SacI (5′) and EcoRI (3′) overhanging restriction sites (Integrated DNA Technologies, Coralville, IA) and hybridized. The hybridized SLN oligomer was inserted into the mVen-N-C1 vector, also digested with SacI and EcoRI. We generated Dysf-mVen-N by amplifying a segment of dysferlin composed of 10 amino acids upstream of the transmembrane domain through its C terminus using primers containing BglII (5′) and EcoRI (3′) and inserting it into the mVen-N-C1 vector. Oligomers encoding the 3× FLAG sequence flanked by EcoRI (5′) and SacII (3′) overhangs were designed and ligated to the C-terminal end of Dysf-mVen-N to generate the Dysf-mVen-N-FLAG construct used in our experiments.
Transfection
COS7 and HEK293 cells (American Type Culture Collection, Manassas, VA) were cultured in DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin in an atmosphere of 10% CO2, 90% air and transiently transfected with cDNA at a concentration of 1 μg/ml and either Lipofectamine 2000 (Invitrogen) or LipoD293 (SignaGen, Gaithersburg, MD), according to manufacturer's protocols. Unless otherwise indicated, cotransfection of two or more cDNAs were carried out using a ratio of 1:2:2 (SERCA1 cDNA/X/Y) for coIP and ATPase assays, and 1:1:1 for AFRET and complementation assays. Empty vectors were cotransfected as controls when appropriate. Cells were seeded to achieve ∼70–80% confluency at time of transfection and incubated for 48 h to allow protein expression. Cells seeded on 35-cm tissue culture plates with glass bottoms (MatTek, Ashland, MA) were washed with Hanks' balanced salt solution + 0.1% BSA and used for AFRET. For preparation of microsomes, cells in 10- or 15-cm tissue culture dishes were washed twice with PBS, detached with a cell scraper, homogenized, and subjected to differential centrifugation (36). The microsomal fraction was used for ATPase measurements. Alternatively, cell extracts were solubilized in a solution containing 0.5% Tween 20 (see below) for coimmunoprecipitation studies.
Coimmunoprecipitation
The coIP experiments were performed as described (37), with preparations of SR vesicles isolated by the method of Eletr and Inesi (38) and Kosk-Kosicka (39) from rabbit skeletal muscle from the back and hind limb (Pel-Freez, Rogers, AR) or crude membrane extracts of COS7 cells prepared as described previously (36, 40). COS7 cells were harvested in PBS at 48 h following transfection, collected by centrifugation, and processed for use or frozen in liquid N2 and stored at −80 °C until needed. Briefly, pellets were homogenized using 30 strokes of a glass Dounce homogenizer in resuspension buffer (0.25 m sucrose, 10 mm Tris-HCl, pH 7.5, 20 mm CaCl2, 3 mm 2-mercaptoethanol, 150 mm KCl) and solubilized with an equal volume of lysis buffer (40 mm HEPES-NaOH, pH 7.5, 300 mm NaCl, 2 mm EDTA, 1% Tween 20). SR vesicles were prepared for coIP by the same methods for resuspension and solubilization. Extracts were mixed with antibody-coated Dynabeads (1 μg of antibody/10 μl of beads) and incubated at 4 °C for 4 h. Following ≥5 washes in washing buffer (20 mm HEPES, pH 7.5, 150 mm NaCl, 1 mm EDTA, 0.5% Tween 20), protein was eluted by boiling the beads in 70 μl of SDS-PAGE sample loading buffer. Proteins in the samples were separated by SDS-PAGE and analyzed via immunoblot as described (41).
Microscopy
Cultures of COS7 cells, transfected as described above, were fixed in PBS containing 4% paraformaldehyde and 4% sucrose for 15 min at room temperature and mounted in Vectashield (Vector Laboratories, Burlingame, CA). Confocal microscopy used a Zeiss 510 META system (Carl Zeiss, Thornwood, NY) equipped with a ×63, 1.4 numerical aperture objective lens. Images were collected by exciting the samples at 458 nm (CFP), 514 nm (YFP), and 543 nm (RFP and mCherry), provided by an argon laser, and collected through 480–520 nm (CFP) BP, 530–600 nm (YFP) BP, and 560 nm (RFP and mCherry) LP filters. Colocalization was assessed by Pearson's coefficients with ImageJ software (rsb.info.nih.gov/ij) and the Just Another Colocalization Plugin (42).
For investigating the interaction between sAnk1, SERCA1, and SLN by AFRET, transfected COS7 cells on glass-bottomed plates were washed with Hanks' balanced salt solution + 0.1% BSA without phenol red and examined with a specially equipped Zeiss AxioObserver microscope (28) with a ×20, 0.75 numerical aperture, dry objective lens. The filters and equations used to collect and analyze the data have been described in detail (28, 43). Briefly, fields were illuminated with polarized light, and the intensity of emitted light was measured in planes parallel and perpendicular to that used for illumination. Emissions were collected for donor, acceptor, and FRET (excite donor, collect acceptor) fluorescence in both planes. ImageJ software was used to stack and align the images (16-bit) and to collect mean intensity values from a minimum of three regions of interest (ROI) per cell. ROIs were excluded if their mean pixel intensity value was <2000 above background. ROIs with directly excited YFP anisotropies less than or equal to FRET anisotropies were excluded from the analysis to minimize the influence of potential YFP homotransfer on heterotransfer FRET measurements. Microsoft Excel was used to calculate anisotropies and FRET, indicated by a non-zero difference between the anisotropy measured for the FRET channel compared with the anisotropy measured for the donor (Δr = rCFP − rFRET).
In more recent studies, we analyzed AFRET pixel-by-pixel, as follows. Parallel and perpendicular polarization images were extracted from raw “dual-view” images and aligned using FIJI software (44). Image data were then loaded into R statistical software package (www.r-project.org)4 using the EBImage package (45). Anisotropy values for each pixel were then calculated. Background, saturated, and low signal-to-noise pixels were excluded from the analysis.
Assay of Ca2+-ATPase activity
ATPase activity was measured in microsomes prepared from transfected COS7 or HEK293 cells with a colorimetric assay to detect inorganic phosphate released during ATP hydrolysis, as described (1, 36).
Statistics
Values are reported as mean ± S.E. All graphs were created with GraphPad Prism 5 software (La Jolla, CA). For the AFRET experiments shown in Fig. 3, each data point represents the average Δr value from all valid ROIs (see under “Microscopy” for criteria). The t tests compared these values to a theoretical mean of zero to determine whether the mean AFRET value was statistically significant. Student's t test was used to compare the amount of sAnk1 present in coIPs generated with or without SLN, with p < 0.05 being considered significant. Data for experiments examining titration of sAnk1 or SLN were fit by linear regression. Significance was determined by a non-zero slope for which p < 0.05. Results of assays of ATPase activity were fit to the equation for an allosteric sigmoidal model (Y = Vmax·[S]h/(K′ + [S]h) (46, 47) from data acquired in four experiments conducted on microsomes from three independent transfections. KCa2+ ([Ca2+]free resulting in half-maximal activation) was calculated using nonlinear regression analysis. Statistics were evaluated with one-way ANOVA with p < 0.05 considered significant.
Author contributions
P. F. D., M. A. R., and R. J. B. designed these experiments and wrote the paper. P. F. D. performed and analyzed experiments shown in Figs. 1, 2, 4, 5, and 8. P. F. D. and A. L. performed the experiments presented in Fig. 7. J. M. designed and prepared fluorescent constructs used for AFRET studies in COS7 cells. M. A. R. contributed expertise in all AFRET studies, proposed the experiments utilizing the half-Venus constructs, and devised the procedures for data analysis. A. L. and M. L. M. prepared samples with half-Venus constructs. M. L. M. collected all AFRET data; the results in Fig. 3 and supplemental Fig. 1 were generated by P. F. D.; and analyses shown in Fig. 8 were done by A. E. M. R. J. B. created the model in Fig. 9 and supplemental Fig. 2.
Supplementary Material
Acknowledgments
SERCA1 and FLAG-SLN cDNA constructs were generous gifts from Dr. David MacLennan (University of Toronto). DS-Red-KDEL was a gift from Dr. Shengyun Fang (University of Maryland, Baltimore). The mVenus-C-N1 and -N-C1 vectors were provided by Dr. Michael Frohmen (State University of New York, Stony Brook).
This work was supported in part by National Institutes of Health Grants RO1 DK077140, R01 HL122827A, and R01 MH111527A (to M. A. R.), National Institutes of Health Grant RO1 AR056330 (to R. J. B.), and The Kahlert Foundation. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Figs. S1 and S2.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- SR
- sarcoplasmic reticulum
- coIP
- coimmunoprecipitation
- AFRET
- anisotropy-based FRET
- SLN
- sarcolipin
- PLN
- phospholamban
- SERCA
- sarco(endo)plasmic reticulum Ca2+-ATPase
- TM
- transmembrane
- sAnk1
- small ankyrin 1
- ANOVA
- analysis of variance
- a.a.
- amino acid
- nSR
- network SR
- mVen
- mVenus
- ROI
- regions of interest
- ER
- endoplasmic reticulum
- IP
- immunoprecipitation
- CFP
- cyan fluorescent protein
- RFP
- red fluorescent protein.
References
- 1. Desmond P. F., Muriel J., Markwardt M. L., Rizzo M. A., and Bloch R. J. (2015) Identification of small ankyrin 1 as a novel sarco(endo)plasmic reticulum Ca2+-ATPase 1 (SERCA1) regulatory protein in skeletal muscle. J. Biol. Chem. 290, 27854–27867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. MacLennan D. H., Asahi M., and Tupling A. R. (2003) The regulation of SERCA-type pumps by phospholamban and sarcolipin. Ann. N.Y. Acad. Sci. 986, 472–480 [DOI] [PubMed] [Google Scholar]
- 3. Periasamy M., and Kalyanasundaram A. (2007) SERCA pump isoforms: their role in calcium transport and disease. Muscle Nerve 35, 430–442 [DOI] [PubMed] [Google Scholar]
- 4. Brini M., and Carafoli E. (2009) Calcium pumps in health and disease. Physiol. Rev. 89, 1341–1378 [DOI] [PubMed] [Google Scholar]
- 5. Díaz M. E., Graham H. K., O'neill S. C., Trafford A. W., and Eisner D. A. (2005) The control of sarcoplasmic reticulum Ca content in cardiac muscle. Cell Calcium 38, 391–396 [DOI] [PubMed] [Google Scholar]
- 6. Schmidt A. G., Zhai J., Carr A. N., Gerst M. J., Lorenz J. N., Pollesello P., Annila A., Hoit B. D., and Kranias E. G. (2002) Structural and functional implications of the phospholamban hinge domain: impaired SR Ca2+ uptake as a primary cause of heart failure. Cardiovasc. Res. 56, 248–259 [DOI] [PubMed] [Google Scholar]
- 7. Vangheluwe P., Tjwa M., Van Den Bergh A., Louch W. E., Beullens M., Dode L., Carmeliet P., Kranias E., Herijgers P., Sipido K. R., Raeymaekers L., and Wuytack F. (2006) A SERCA2 pump with an increased Ca2+ affinity can lead to severe cardiac hypertrophy, stress intolerance and reduced life span. J. Mol. Cell. Cardiol. 41, 308–317 [DOI] [PubMed] [Google Scholar]
- 8. Stammers A. N., Susser S. E., Hamm N. C., Hlynsky M. W., Kimber D. E., Kehler D. S., and Duhamel T. A. (2015) The regulation of sarco(endo)plasmic reticulum calcium-ATPases (SERCA). Can. J. Physiol. Pharmacol. 93, 843–854 [DOI] [PubMed] [Google Scholar]
- 9. Bhupathy P., Babu G. J., and Periasamy M. (2007) Sarcolipin and phospholamban as regulators of cardiac sarcoplasmic reticulum Ca2+ ATPase. J. Mol. Cell. Cardiol. 42, 903–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Anderson D. M., Anderson K. M., Chang C. L., Makarewich C. A., Nelson B. R., McAnally J. R., Kasaragod P., Shelton J. M., Liou J., Bassel-Duby R., and Olson E. N. (2015) A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 160, 595–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Anderson D. M., Makarewich C. A., Anderson K. M., Shelton J. M., Bezprozvannaya S., Bassel-Duby R., and Olson E. N. (2016) Widespread control of calcium signaling by a family of SERCA-inhibiting micropeptides. Sci. Signal. 9, ra119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nelson B. R., Makarewich C. A., Anderson D. M., Winders B. R., Troupes C. D., Wu F., Reese A. L., McAnally J. R., Chen X., Kavalali E. T., Cannon S. C., Houser S. R., Bassel-Duby R., and Olson E. N. (2016) A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 351, 271–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou D., Birkenmeier C. S., Williams M. W., Sharp J. J., Barker J. E., and Bloch R. J. (1997) Small, membrane-bound, alternatively spliced forms of ankyrin 1 associated with the sarcoplasmic reticulum of mammalian skeletal muscle. J. Cell Biol. 136, 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Birkenmeier C. S., Sharp J. J., Gifford E. J., Deveau S. A., and Barker J. E. (1998) An alternative first exon in the distal end of the erythroid ankyrin gene leads to production of a small isoform containing an NH2-terminal membrane anchor. Genomics 50, 79–88 [DOI] [PubMed] [Google Scholar]
- 15. Birkenmeier C. S., White R. A., Peters L. L., Hall E. J., Lux S. E., and Barker J. E. (1993) Complex patterns of sequence variation and multiple 5′ and 3′ ends are found among transcripts of the erythroid ankyrin gene. J. Biol. Chem. 268, 9533–9540 [PubMed] [Google Scholar]
- 16. Armani A., Galli S., Giacomello E., Bagnato P., Barone V., Rossi D., and Sorrentino V. (2006) Molecular interactions with obscurin are involved in the localization of muscle-specific small ankyrin1 isoforms to subcompartments of the sarcoplasmic reticulum. Exp. Cell Res. 312, 3546–3558 [DOI] [PubMed] [Google Scholar]
- 17. Ackermann M. A., Ziman A. P., Strong J., Zhang Y., Hartford A. K., Ward C. W., Randall W. R., Kontrogianni-Konstantopoulos A., and Bloch R. J. (2011) Integrity of the network sarcoplasmic reticulum in skeletal muscle requires small ankyrin 1. J. Cell Sci. 124, 3619–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gagelin C., Constantin B., Deprette C., Ludosky M. A., Recouvreur M., Cartaud J., Cognard C., Raymond G., and Kordeli E. (2002) Identification of Ank(G107), a muscle-specific ankyrin-G isoform. J. Biol. Chem. 277, 12978–12987 [DOI] [PubMed] [Google Scholar]
- 19. Kontrogianni-Konstantopoulos A., Jones E. M., Van Rossum D. B., and Bloch R. J. (2003) Obscurin is a ligand for small ankyrin 1 in skeletal muscle. Mol. Biol. Cell 14, 1138–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kontrogianni-Konstantopoulos A., and Bloch R. J. (2003) The hydrophilic domain of small ankyrin-1 interacts with the two N-terminal immunoglobulin domains of titin. J. Biol. Chem. 278, 3985–3991 [DOI] [PubMed] [Google Scholar]
- 21. Giacomello E., Quarta M., Paolini C., Squecco R., Fusco P., Toniolo L., Blaauw B., Formoso L., Rossi D., Birkenmeier C., Peters L. L., Francini F., Protasi F., Reggiani C., and Sorrentino V. (2015) Deletion of small ankyrin 1 (sAnk1) isoforms results in structural and functional alterations in aging skeletal muscle fibers. Am. J. Physiol. Cell Physiol. 308, C123–C138 [DOI] [PubMed] [Google Scholar]
- 22. Porter N. C., Resneck W. G., O'Neill A., Van Rossum D. B., Stone M. R., and Bloch R. J. (2005) Association of small ankyrin 1 with the sarcoplasmic reticulum. Mol. Membr. Biol. 22, 421–432 [DOI] [PubMed] [Google Scholar]
- 23. Autry J. M., Rubin J. E., Pietrini S. D., Winters D. L., Robia S. L., and Thomas D. D. (2011) Oligomeric interactions of sarcolipin and the Ca-ATPase. J. Biol. Chem. 286, 31697–31706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Morita T., Hussain D., Asahi M., Tsuda T., Kurzydlowski K., Toyoshima C., and Maclennan D. H. (2008) Interaction sites among phospholamban, sarcolipin, and the sarco(endo)plasmic reticulum Ca2+-ATPase. Biochem. Biophys. Res. Commun. 369, 188–194 [DOI] [PubMed] [Google Scholar]
- 25. Asahi M., Kurzydlowski K., Tada M., and MacLennan D. H. (2002) Sarcolipin inhibits polymerization of phospholamban to induce superinhibition of sarco(endo)plasmic reticulum Ca2+-ATPases (SERCAs). J. Biol. Chem. 277, 26725–26728 [DOI] [PubMed] [Google Scholar]
- 26. Hellstern S., Pegoraro S., Karim C. B., Lustig A., Thomas D. D., Moroder L., and Engel J. (2001) Sarcolipin, the shorter homologue of phospholamban, forms oligomeric structures in detergent micelles and in liposomes. J. Biol. Chem. 276, 30845–30852 [DOI] [PubMed] [Google Scholar]
- 27. Markwardt M. L., Kremers G. J., Kraft C. A., Ray K., Cranfill P. J., Wilson K. A., Day R. N., Wachter R. M., Davidson M. W., and Rizzo M. A. (2011) An improved cerulean fluorescent protein with enhanced brightness and reduced reversible photoswitching. PLoS ONE 6, e17896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Piston D. W., and Rizzo M. A. (2008) FRET by fluorescence polarization microscopy. Methods Cell Biol. 85, 415–430 [DOI] [PubMed] [Google Scholar]
- 29. Rizzo M. A., and Piston D. W. (2005) High-contrast imaging of fluorescent protein FRET by fluorescence polarization microscopy. Biophys. J. 88, L14–L16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kerppola T. K. (2008) Biomolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 37, 465–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rizzo M. A., Springer G., Segawa K., Zipfel W. R., and Piston D. W. (2006) Optimization of pairings and detection conditions for measurement of FRET between cyan and yellow fluorescent proteins. Microsc. Microanal. 12, 238–254 [DOI] [PubMed] [Google Scholar]
- 32. Fajardo V. A., Bombardier E., Vigna C., Devji T., Bloemberg D., Gamu D., Gramolini A. O., Quadrilatero J., and Tupling A. R. (2013) Co-expression of SERCA isoforms, phospholamban and sarcolipin in human skeletal muscle fibers. PLoS ONE 8, e84304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Toyoshima C., Asahi M., Sugita Y., Khanna R., Tsuda T., and MacLennan D. H. (2003) Modeling of the inhibitory interaction of phospholamban with the Ca2+ ATPase. Proc. Natl. Acad. Sci. U.S.A. 100, 467–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Asahi M., Sugita Y., Kurzydlowski K., De Leon S., Tada M., Toyoshima C., and MacLennan D. H. (2003) Sarcolipin regulates sarco(endo)plasmic reticulum Ca2+-ATPase (SERCA) by binding to transmembrane helices alone or in association with phospholamban. Proc. Natl. Acad. Sci. U.S.A. 100, 5040–5045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schneider J. S., Shanmugam M., Gonzalez J. P., Lopez H., Gordan R., Fraidenraich D., and Babu G. J. (2013) Increased sarcolipin expression and decreased sarco(endo)plasmic reticulum Ca2+ uptake in skeletal muscles of mouse models of Duchenne muscular dystrophy. J. Muscle Res. Cell Motil. 34, 349–356 [DOI] [PubMed] [Google Scholar]
- 36. Fu M. H., and Tupling A. R. (2009) Protective effects of Hsp70 on the structure and function of SERCA2a expressed in HEK-293 cells during heat stress. Am. J. Physiol. Heart Circ. Physiol. 296, H1175–H1183 [DOI] [PubMed] [Google Scholar]
- 37. Asahi M., Kimura Y., Kurzydlowski K., Tada M., and MacLennan D. H. (1999) Transmembrane helix M6 in sarco(endo)plasmic reticulum Ca2+-ATPase forms a functional interaction site with phospholamban. Evidence for physical interactions at other sites. J. Biol. Chem. 274, 32855–32862 [DOI] [PubMed] [Google Scholar]
- 38. Eletr S., and Inesi G. (1972) Phase changes in the lipid moieties of sarcoplasmic reticulum membranes induced by temperature and protein conformational changes. Biochim. Biophys. Acta 290, 178–185 [DOI] [PubMed] [Google Scholar]
- 39. Kosk-Kosicka D. (2013) Measurement of Ca2+-ATPase activity (in PMCA and SERCA1). Methods Mol. Biol. 937, 343–356 [DOI] [PubMed] [Google Scholar]
- 40. Maruyama K., and MacLennan D. H. (1988) Mutation of aspartic acid-351, lysine-352, and lysine-515 alters the Ca2+ transport activity of the Ca2+-ATPase expressed in COS-1 cells. Proc. Natl. Acad. Sci. U.S.A. 85, 3314–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lencesova L., O'Neill A., Resneck W. G., Bloch R. J., and Blaustein M. P. (2004) Plasma membrane-cytoskeleton-endoplasmic reticulum complexes in neurons and astrocytes. J. Biol. Chem. 279, 2885–2893 [DOI] [PubMed] [Google Scholar]
- 42. Bolte S., and Cordelières F. P. (2006) A guided tour into subcellular co-localization analysis in light microscopy. J. Microsc. 224, 213–232 [DOI] [PubMed] [Google Scholar]
- 43. Gade P., Ramachandran G., Maachani U. B., Rizzo M. A., Okada T., Prywes R., Cross A. S., Mori K., and Kalvakolanu D. V. (2012) An IFN-γ-stimulated ATF6-C/EBP-β-signaling pathway critical for the expression of death associated protein kinase 1 and induction of autophagy. Proc. Natl. Acad. Sci. U.S.A. 109, 10316–10321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., and Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pau G., Fuchs F., Sklyar O., Boutros M., and Huber W. (2010) EBImage–an R package for image processing with applications to cellular phenotypes. Bioinformatics 26, 979–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bombardier E., Smith I. C., Vigna C., Fajardo V. A., and Tupling A. R. (2013) Ablation of sarcolipin decreases the energy requirements for Ca2+ transport by sarco(endo)plasmic reticulum Ca2+-ATPases in resting skeletal muscle. FEBS Lett. 587, 1687–1692 [DOI] [PubMed] [Google Scholar]
- 47. Sahoo S. K., Shaikh S. A., Sopariwala D. H., Bal N. C., Bruhn D. S., Kopec W., Khandelia H., and Periasamy M. (2015) The N terminus of sarcolipin plays an important role in uncoupling sarco-endoplasmic reticulum Ca2+ ATPase (SERCA) ATP hydrolysis from Ca2+ transport. J. Biol. Chem. 290, 14057–14067 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.