Abstract
Mutations in the thymidine kinase (TK) gene of herpes simplex virus (HSV) may confer resistance to acyclovir (ACV). Because of the high genetic polymorphism of this gene, discriminating between mutations related to resistance and mutations related to gene polymorphism can be difficult, especially when no sensitive strain has been previously isolated from the same patient. To assess the role of the mutations located at codons 51, 77, 83, and 175, previously detected in HSV-1 clinical isolates (F. Morfin, G. Souillet, K. Bilger, T. Ooka, M. Aymard, and D. Thouvenot, J. Infect. Dis. 182:290-293, 2000), in the acquisition of resistance to ACV, four mutants with site-directed mutations at these respective codons were constructed. The enzymatic activity of the proteins, produced using both a reticulocyte lysate system and a bacterial system, was evaluated using [3H]thymidine as substrate. This site-directed mutagenesis revealed that mutations at codons 51, 83, and 175 induce a loss of HSV-1 TK activity and are thus clearly involved in the acquisition of resistance to ACV. On the other hand, the mutation at codon 77 does not affect enzyme activity.
Acyclovir (ACV) is the accepted first-line treatment for herpes simplex virus (HSV) infections. This antiviral drug is a guanosine analog that must be phosphorylated to its triphosphate form to become active. The first phosphorylation is completed by a thymidine kinase (TK) encoded by the virus, and the two further phosphorylations are performed by cellular thymidylate kinases. The triphosphate form of ACV, a competitive inhibitor of viral DNA polymerase, is incorporated in nascent new viral DNA to block further viral replication.
Since it was first used, strains resistant to ACV have been isolated with a prevalence of less than 1% in immunocompetent patients (6, 9) but 5% of in immunocompromised patients (5, 6) and 25% in allogenic bone marrow transplant patients (4).
HSV resistance to ACV has been associated with mutations occurring either in the viral TK gene or the viral DNA polymerase gene. Three mechanisms of resistance have been proposed: a loss of TK activity (TK-deficient virus), an alteration of TK substrate specificity (TK-altered virus), and/or an alteration of DNA polymerase activity (15). In 95% of the strains, resistance is associated with TK mutations.
HSV TK is a 376-amino-acid protein encoded by the UL23 gene. It has an ATP binding site (codons 51 to 63) and a nucleoside binding site (codons 168 to 176), and there are six conserved regions among Herpesviridae TKs (1). Mutations related to resistance are nucleotide insertions, deletions, or substitutions. Nucleotide insertions or deletions have been reported in 50% of the cases described so far (11, 17). These cause a frameshift, resulting in the synthesis of a nonfunctional truncated TK. Most of these mutations occur in guanosine (G) or cytidine (C) homopolymer repeats, which are thought to be hot spots for mutations (11, 20). The two longest homopolymers, composed of 7 G's (codons 144 to 146) and of 6 C's (codons 183 to 185), are the site of the most frequent mutations reported in ACV-resistant clinical isolates (11, 21). Resistance in other strains is associated with substitutions usually located in the conserved regions of the TK gene, the most frequent being the substitution of arginine at codon 176 of HSV type I (HSV-1) TK and at codon 177 of HSV type 2 (HSV-2) TK and the substitution of cysteine at codon 336 of HSV-1 TK and at codon 337 of HSV-2 TK (12). In addition to these mutations related to ACV resistance, genetic characterization of sensitive strains revealed a large degree of polymorphism in HSV TK (3, 14, 17). These mutations unrelated to resistance are located throughout the gene but mainly outside of the active or conserved sites. Because a large number of substitutions have been found in clinical isolates of HSV, it may be difficult to differentiate between mutations associated with gene polymorphism and those associated with drug resistance when no previous isolate from the same patient is available for sequence comparison.
Our laboratory has previously carried out the phenotypic and genotypic characterization of 20 clinical ACV-susceptible or resistant HSV-1 isolates (17). This showed 19 amino acid substitutions related to TK gene polymorphism. One of these, D77N, even if detected in a resistant isolate, was likely to be associated with polymorphism which did not affect TK activity, because this strain also showed a nucleotide insertion at codon 146 which may account for ACV resistance. Three other substitutions, R51W, E83K, and A175V, not previously reported, have been detected in ACV-resistant isolates from patients from whom no sensitive strain had been isolated previously. It has been assumed that they were likely to be associated with resistance because all were located in conserved sites.
The objective of the work presented here was to assess the consequence of mutations at codons 51, 77, 83, and 175 in the acquisition of resistance to ACV. Four mutants with site-directed mutations at codons R51W, D77N, E83K, and A175V of the TK gene of an ACV-sensitive HSV-1 strain (KOS) were constructed. Recombinant proteins were synthesized using two different systems: a reticulocyte lysate in vitro system and a bacterial system. The enzymatic activity of the proteins obtained with the two systems was evaluated using [3H]thymidine as the substrate. Our results demonstrated that mutations R51W, E83K, and A175V induced a loss of TK activity and were thus clearly implicated in the acquisition of resistance to ACV. On the other hand, the mutation D77N did not impair TK activity and was likely to be related to simple TK polymorphism.
MATERIALS AND METHODS
Reference strains.
KOS is a TK-positive and ACV-sensitive reference HSV-1 strain (ATCC VR 1493). DM21 is a TK-deficient and ACV-resistant reference HSV-1 strain with a 816-bp deletion between codons 98 and 370 in its TK gene (8).
TK protein production by the reticulocyte lysate system. (i) Construction of plasmids pGEM-TK KOS and pGEM-TK DM21.
The KOS and DM21 strains were propagated in MRC-5 cells in Eagle minimal essential medium (Cambrex) supplemented with 2% fetal calf serum. Infected cells were collected when the cytopathic effect reached 75%, and total DNA was extracted using a standard phenol-chloroform method (18).
The viral TK gene was amplified using 1 μg of extracted DNA and the GC-rich PCR system kit (Roche), with the forward primer TKf (5′-GATCTTGGTGGCGTGAAACTCC-3′) and the reverse primer TKr (5′-GGTTCCTTCCGGTATTGTCTCC-3′). Amplification conditions included an initial denaturation step of 10 min at 95°C followed by 35 cycles of 20 s at 95°C, 30 s at 55°C, and 1 min at 75°C, with a final extension step of 5 min at 75°C. PCR products were separated by agarose gel electrophoresis and purified with the GFX Gel Band purification kit (Amersham). Purified PCR products were cloned into the pGEM-T Easy vector under the control of the T7 promoter (Promega) as described by Maniatis et al. (16). The insert was checked by restriction analysis with PstI. pGEM-TK KOS was used as a positive control and as a matrix to construct mutants. pGEM-TK DM21 was the negative control.
(ii) Construction of mutants pGEM-TKm51, pGEM-TKm77, pGEM-TKm83, and pGEM-TKm175 by site-directed mutagenesis.
Site-directed mutagenesis was performed as described by Higuchi (13). The modified internal forward and reverse primers used in primary PCRs are shown in Table 1. The proofreading Pfx polymerase (Invitrogen) was used for these PCRs. Amplification conditions included an initial denaturation step of 2 min at 94°C followed by 35 cycles of 15 s at 94°C, 30 s at 55°C, and 2 min at 68°C, with a final extension step of 5 min at 68°C. The PCR products were extracted from an agarose gel with the GFX Gel Band Purification kit. The secondary PCR was performed with the GC rich PCR system kit as described above, using TKf and TKr as primers. The cloned fragments were sequenced in their entirety to check for any mutations introduced during PCR (Sequentia, Clermont-Ferrand, France).
TABLE 1.
Modified internal primers used for site-directed mutagenesis
| Mutated codon | Forward primer
|
Reverse primer
|
||
|---|---|---|---|---|
| Name | Sequencea | Name | Sequencea | |
| 51 | TK51f | 5′-TACTGTGGGTTTATATAGACGGT | TK51r | 5′-TAAACCCACAGTAGCGTGGGCAT |
| 77 | TK77f | 5′-GCGACAATATCGTCTACGTACCC | TK77r | 5′-ACGATATTGTCGCGCGAACCCAG |
| 83 | TK83f | 5′-TACCCAAGCCGATGACTTACTGG | TK83r | 5′-ATCGGCTTGGGTACGTAGACGAT |
| 146 | TK146f | 5′-GGGGGGAGGCTGGGAGCTCACAT | TK146r | 5′-CCAGCCTCCCCCCCGATATGA |
| 175 | TK175f | 5′-CCGGCCGTGCGATACCTTATGGG | TK175r | 5′-TATCGCACGGCCGGGTAGCACAG |
The mutated nucleotide is shown in bold type for each sequence.
(iii) In vitro TK protein production.
The TK protein was synthesized from constructed plasmids using the TNT-coupled reticulocyte lysate system (Promega). Briefly, a reaction mixture with a total volume of 50 μl containing 1 μg of plasmid DNA, 1 μl of T7 RNA polymerase, 25 μl of rabbit reticulocytes, 2 μl of amino acid mixture, and 40 u of RNase inhibitor (Recombinant RNasin, Promega) was prepared. This was incubated for 2 h at 30°C. The enzymatic activity of the newly synthesized proteins was analyzed without any purification. Reticulocytes without any plasmid DNA were analyzed to check for the absence of any phosphorylating activity.
To estimate the quantity of synthesized protein, transcription-translation of the TK gene was performed in parallel with a [35S]methionine label by adding 2 μl of Redivue l-[35S]methionine (1,000 Ci/mmol; Amersham) to the mix described above. A sodium dodecyl sulfate-(SDS)-polyacrylamide gel was run, dried, and placed on a radioactivity tape, which was scanned after 48 h with a phosphorimager (Bio-Rad). The luciferase protein (∼61 kDa) was used as a positive control.
TK protein production by bacterial systems. (i) Construction of plasmids pT7.7-KOS, pT7.7-TKm51, pT7.7-TKm77, pT7.7-TKm83, and pT7.7-TKm175.
The previously constructed plasmids pGEM-TK KOS, pGEM-TKm51, pGEM-TKm77, pGEM-TKm83, and pGEM-TKm175 were used as matrix for the construction of pT7.7-KOS and the four pT7.7-TK mutants. The TK gene was amplified by PCR using the forward primer BTK1 containing the BamHI site (underlined) 5′-CAGGATCCATGGCTTCGTACCCCTGCCAT-3′ and the reverse primer HTK2 containing the HindIII site 5′-TGAAGCTTTCAGTTAGCCTCCCCCATCTC-3′. The amplified fragment was cut twice using BamHI and HindIII and ligated into the corresponding sites of pT7.7 which includes six histidine residues that will be fused at the N terminus of the protein (23). This construct was used to transform Escherichia coli BL21(DE3) cells (Invitrogen).
(ii) TK protein production.
E. coli BL21(DE3) cells harboring plasmid pT7.7-KOS or its mutant derivatives were grown in Luria-Bertani medium to mid-log phase and induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Recombinant proteins were allowed to accumulate for 3 h at 37°C. The cells were harvested and lysed for 30 min at 4°C in 20 mM phosphate buffer (pH 7.4) containing lysozyme (1 mg ml−1). The cell lysates were then incubated with 1% Triton-X-100 for 10 min at 4°C and sonicated for three periods of 30 s. Insoluble material was removed by centrifugation (15 min at 25,000 × g). Imidazole was added to the supernatant to a final concentration of 10 mM.
His-tagged proteins were purified by using standard nickel affinity chromatography. Briefly, Ni-nitroltriacetate Superflow resin (Qiagen) was incubated with the supernatant for 30 min at 4°C. The resin was washed three times with 20 mM phosphate buffer containing 20 mM imidazole. The His-tagged TK was eluted in 20 mM phosphate buffer containing 250 mM imidazole. Fractions of each purification step were analyzed on SDS-12% polyacrylamide gels. Total protein concentration was measured using the Bio-Rad Dc Protein Assay (Bio-Rad).
(iii) Western blot analysis.
Identification of the TK protein was performed by Western blot analysis using polyclonal rabbit antibodies directed against HSV-1 proteins (kindly provided by William C. Summers, Yale University). The proteins were denatured at 100°C for 5 min, electrophoresed at 45 mA for 5 h through a 10% acrylamide gel, and then blotted onto a nitrocellulose membrane by using an electroblotting transfer apparatus (Pharmacia Biotech Inc., San Francisco, Calif.). Antibody incubations and detections were performed with the Supersignal West Pico chemiluminescent substrate kit (Pierce). Briefly, the membrane was saturated with 5% milk blocking solution for 1 h at room temperature and then incubated for 1 h at room temperature with a primary antibody diluted 1:2,500. It was then washed three times in wash buffer (phosphate-buffered saline, 1% Tween) and incubated with a secondary antibody diluted 1:2,000 (anti-rabbit immunoglobulin G-peroxidase [Sigma]). The membrane was washed three more times with phosphate-buffered saline, and a chemiluminescent substrate was then applied to the membrane for 1 min prior to the exposure of the membrane to X-ray film.
TK enzymatic activity.
The enzymatic activity of TK was determined by using [3H]thymidine (91 Ci mmol−1, 1 mCi ml−1 [Amersham]) as previously described (18). For the reticulocyte lysate system, the product of the in vitro transcription-translation step (50 μl) was incubated at 37°C with an identical volume of substrate medium containing 150 mM phosphate buffer (pH 7.5), 20 mM ATP, 20 mM MgCl2, 40 mM KCl, 1 mM dithiothreitol, 10 mM NaF, 10 μM cold thymidine, and 2.5 μCi of [3H]thymidine. For TK produced by the bacterial system, 1 μg of purified His-tagged TK protein was incubated with 50 μl of substrate medium at 37°C. The phosphorylation levels of thymidine at 15, 30, and 60 min were determined by spotting 20 μl of the reaction mixture onto a DEAE paper disk (DE81; Whatman). The disks were subsequently washed three times in 1 mM ammonium formate, dried in ethanol, and counted by scintillation (UltimaGold MV; Packard).
RESULTS
TK production.
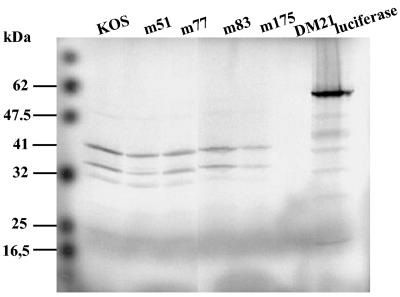
Figure 1 shows the [35S]TK protein produced by the reticulocyte system for KOS, mutant R51W, mutant D77N, mutant E83K, mutant A175V, and mutant DM21. The same volume of reaction mixture (15 μl of 50 μl) was loaded onto polyacrylamide gel. The mutated TK protein was visualized by autoradiography; TK produced by pGEM-TK KOS was used as a control. The intensity of each band was similar except for A175V, which showed a slightly lower intensity than the others. The TK produced with the pGEM-TK DM21 vector was not detected by autoradiography at 48 h since this strain contains an 816-bp deletion in its TK gene, leading to the production of a smaller (∼14-kDa) TK protein. After a long exposure (4 weeks), a very weak band could be visualized at the expected size (data not shown). Full-length TK proteins were obtained with all mutants tested, in a monomeric form of ∼41 kDa. Two smaller peptides of ∼36 and ∼34 kDa were detected. These are likely to be due to the presence of ATG start codons at positions 45 and 59, leading to an alternative translation (22). We were able to use TK produced by this system to measure its activity.
FIG. 1.
[35S]methionine labeling of TK proteins produced using the reticulocyte lysate system. KOS, m51, m77, m83, m175, and DM21 are recombinant proteins of the respective mutants. Luciferase is the positive control of the reaction. A 15-μl sample of each product of the transcription-translation reaction was electrophoresed on an SDS-12% polyacrylamide gel and autoradiographed after 48 h.
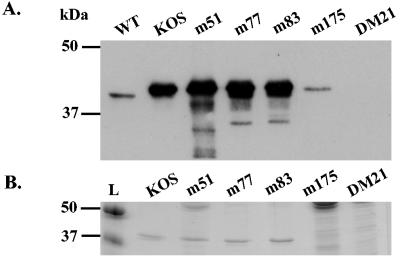
Using the bacterial system, we could obtain about 2.2 mg of each TK starting from 500 ml of culture. Two identical polyacrylamide gels were prepared, and 0.5 mg of each TK was loaded onto each gel (Fig. 2). After electrophoresis, one was stained with Coomassie blue (Fig. 2B) and the other was incubated with antibody against HSV TK (Fig. 2A). Western blot analysis with anti-TK HSV-1 polyclonal antibodies revealed that the recombinant TK proteins produced by this system had antigenic sites and were of the expected molecular mass. However, all the recombinant TKs produced in this system had a higher molecular mass (∼42 kDa) than the wild-type protein did (∼41 kDa). This extra mass is likely to be from the additional tag sequence. The bacterial system allowed the production of proteins for KOS, mutant R51W, mutant D77N, and mutant E83K but not for mutants A175V and DM21.
FIG. 2.
TK proteins produced using the bacterial system. (A) Western blot analysis of TK proteins. A 0.5-μg sample of each purified TK was loaded onto an SDS-10% polyacrylamide gel, transferred, and reacted with polyclonal anti HSV1-TK. (B) Standardization of the quantity of recombinant proteins. A 0.5-μg sample of each purified TK was electrophoresed on an SDS-12% polyacrylamide gel. The gel was stained with Coomassie blue. WT indicates TK from wild-type HSV-1 is the positive control; KOS, m51, m77, m83, m175, and DM21 are recombinant proteins of the respective mutants; DM21 is the negative control; L indicates the DNA ladder.
TK activity.
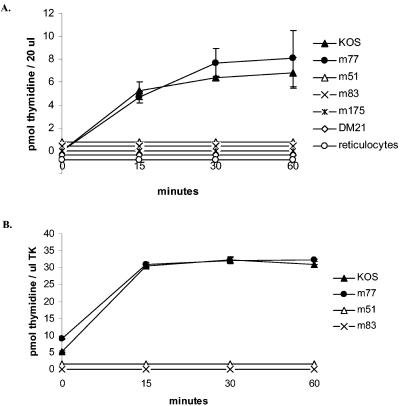
Enzyme activities were evaluated by assaying the transformation of radioactive thymidine to thymidine monophosphate. Adsorption of the reaction product onto a DEAE-cellulose paper after 0, 15, 30, and 60 min of incubation allowed us to separate the phosphorylated thymidine from the nonphosphorylated substrate. This technique reflected the capacity of the TK to phosphorylate the substrate. Previous data from experiments with extracts of cells infected with the KOS reference strain showed that the enzymatic activity measured after 15, 30, and 60 min of incubation increased linearly (18).
The enzymatic activities of the proteins produced by the reticulocyte system at the indicated times are shown in Fig. 3A. Means and standard deviations were calculated from three independent assays performed for each mutant. The results showed that mutant D77N had a phosphorylating activity of 8.07 ± 1.38 pmol of thymidine/h/20 μl of reactional mixture. This activity was similar to KOS activity (6.81 ± 2.46 pmol of thymidine/h/20 μl of reactional mixture). Moreover, mutant D77N had the same phosphorylating initial velocity at 15 min as KOS (19 and 21 pmol of thymidine/h/20 μl of reactional mixture, respectively), showing that this mutant had a functional TK activity. Mutations R51W, E83K, and A175V completely abolished the phosphorylating activity of the respective mutant proteins, defining them as TK negative.
FIG. 3.
Enzymatic activities of TK proteins. (A) TK proteins produced using the reticulocyte lysate system. (B) TK proteins produced using the bacterial system. KOS is the positive control, and DM21 is the negative control. m51, m77, m83, m175, and DM21 are recombinant proteins of the respective mutants. Reticulocytes are the control of residual activity (reaction without any plasmid DNA to transcribe). Means and standard deviations (error bars) are calculated from three independent experiments performed for each mutant.
The activities of the proteins produced using the bacterial system are shown in Fig. 3B. Before testing for enzymatic activity, the quantity of protein was normalized to 0.5 μg for each TK (Fig. 2B). As observed with the TK produced by the reticulocyte lysate system, mutant D77N maintained a conserved phosphorylating activity of 32.22 ± 0.33 pmol of thymidine/μg of TK/h, which is equivalent to the phosphorylating activity of KOS (30.89 ± 0.03 pmol of thymidine/μg of TK/h). The phosphorylating activity of the TK was abolished by mutations R51W and E83K. These data agree with the results obtained with the reticulocyte system.
DISCUSSION
Resistance to ACV is currently detected by phenotypic assays to determine the concentration of antiviral drug that inhibits 50% of viral replication in vitro. Phenotypic methods require virus isolation in cell culture, which is time-consuming and may delay any modification of antiviral treatment based on in vitro susceptibility. Direct sequencing of the HSV TK gene from a biological sample could be useful in detecting antiviral drug resistance more rapidly. Nevertheless, the relationship between a mutation discovered in the HSV TK gene and resistance to ACV is frequently difficult to establish because of the heterogeneity of mutations found in this gene. Only some of these mutations are responsible for acquisition of resistance to ACV; the mutations associated with TK gene polymorphism do not affect TK activity (14). The interpretation of HSV TK gene-sequencing results is consequently difficult, especially when a sensitive strain has not been previously isolated from the same patient. A database of HSV TK mutations associated with either ACV resistance or TK polymorphism is essential for the interpretation of sequencing results. Using such a database, rapid molecular detection of resistance can be performed in 24 to 48 h, allowing early adaptation of antiviral treatment.
This study permitted us to identify mutations at codons 51 (R51W), 83 (E83K), and 175 (A175V) of HSV TK gene as responsible for the suppression of TK phosphorylating activity and resistance to ACV. Mutations at codon 77 (D77N) do not change TK enzymatic activity and probably reflect simple TK gene polymorphism.
Recombinant proteins have been produced either in a reticulocyte lysate in vitro system or in bacteria. The two techniques allowed studies of the enzymatic activity of these proteins and differentiation of mutations related to simple polymorphism from those related to resistance. The transcription-translation assay using the reticulocyte lysate system is a simple and quick, although expensive, way to produce functional recombinant proteins (22). The enzymatic activity determined with this system was at a similar level in several independent repeat assays, and [35S]methionine labeling is a valid way to estimate protein production. The bacterial system, which produces larger amounts of recombinant proteins, allowed standardization of protein quantity before use in the enzymatic assay. However, there are disadvantages with this system, since some specific mutant proteins were found as inclusion bodies. In our study, mutant A175V and DM21 proteins were not produced in the bacterial system but were produced in the reticulocyte one. To explore this further, the solubility of these specific proteins could be assessed in eucaryotic expression system such as Pichia pasteuris yeast (19). Since all the proteins could be produced in the reticulocyte lysate system, it would be an efficient screening method to discriminate TK-positive mutants (mutations related to simple polymorphism) from TK-negative mutants (mutations related to resistance). For any TK-negative mutants, the protein could also be expressed in the bacterial system, since it allows standardization of protein quantity, to confirm that this mutation is related to resistance to ACV.
Mutations located on conserved regions of the TK gene are most often involved in resistance to ACV. At codon 51, which is located in the ATP binding site of the enzyme (1), replacing the gene encoding arginine with that encoding tryptophan, a hydrophobic residue, suppresses TK-phosphorylating activity. Because of their positive environment, arginine residues are known to be involved in amino acid interactions by making connections with carboxylic groups of other amino acids (10). It has already been shown that P57H, D59P, and K62N mutations in this ATP binding site are associated with resistance (2, 11, 21).
Our data show that mutation E83K, also located in a conserved region of the TK gene, is implicated in abolishing TK activity. The amino acid at codon 83 is analogous to the glutamine at position 48 in the TK of varicella-zoster virus, which has already been detected in ACV-resistant varicella-zoster virus isolates (18).
Mutation A175V is located in the nucleoside binding site. Using the reticulocyte system, we assessed the role of this mutation at codon 175 in ACV resistance. In this active sequence of 9 amino acids, two other substitutions in the HSV-1 TK gene, A168T and R176Q, have already been associated with resistance to ACV (7).
Mutation D77N, located in an unconserved region, has already been detected in an ACV-resistant clinical strain (17). However, the presence in this strain of a second mutation (the addition of one G at codon 146) suggests that the mutation at codon 77 would be likely to be associated with simple TK polymorphism (17). Our work confirmed that TK activity is not impaired by such a substitution.
Our work confirms the role played by some amino acids located in the ATP and nucleoside binding sites and in other TK conserved regions. These amino acids are strongly involved in TK phosphorylating activity. On the other hand, substitutions located outside the TK-conserved sites are likely to be associated with TK gene polymorphism, but their role needs, in each case, to be confirmed by site-directed mutagenesis.
To characterize enzymatic activity, the reticulocyte system is convenient, easy, and quick. Production of recombinant TK in the bacterial system could be used as a confirmation assay. These results have to be extended by similar analyses of other mutations in the TK gene associated with ACV-resistant HSV and determinations of whether they are related to resistance or to simple polymorphism. Such data will be useful in interpreting TK gene-sequencing results and in promoting the use of rapid molecular biology tests to detect resistance. The availability of such tests will help clinicians to manage antiviral treatment of HSV infections.
Acknowledgments
We thank Christel Matias for excellent technical assistance.
REFERENCES
- 1.Balasubramaniam, N. K., V. Veerisetty, and G. A. Gentry. 1990. Herpesviral deoxythymidine kinases contain a site analogous to the phosphoryl-binding arginine-rich region of porcine adenylate kinase; comparison of secondary structure predictions and conservation. J. Gen. Virol. 71:2979-2987. [DOI] [PubMed] [Google Scholar]
- 2.Bestman-Smith, J., L. Schmit, B. Papadopoulou, and G. Boivin. 2001. Highly reliable heterologous system for evaluating resistance of clinical herpes simplex virus isolates to nucleoside analogues. J. Virol. 75:3105-3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodaghi, B., C. Mougin, S. Michelson, H. Agut, P. Dighiero, H. Offret, and E. Frau. 2000. Acyclovir-resistant bilateral keratitis associated with mutations in the HSV-1 thymidine kinase gene. Exp. Eye Res. 71:353-359. [DOI] [PubMed] [Google Scholar]
- 4.Chakrabarti, S., D. Pillay, D. Ratcliffe, P. A. Cane, K. E. Collingham, and D. W. Milligan. 2000. Resistance to antiviral drugs in herpes simplex virus infections among allogeneic stem cell transplant recipients: risk factors and prognostic significance. J. Infect. Dis. 181:2055-2058. [DOI] [PubMed] [Google Scholar]
- 5.Christophers, J., J. Clayton, J. Craske, R. Ward, P. Collins, M. Trowbridge, and G. Darby. 1998. Survey of resistance of herpes simplex virus to acyclovir in Northwest England. Antimicrob. Agents Chemother. 42:868-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danve-Scatanek, C., M. Aymard, and D. Thouvenot, et al. 2004. A surveillance network of the herpes simplex virus resistance to antiviral drugs: a three year follow-up. J. Clin. Microbiol. 42:242-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darby, G., B. A. Larder, and M. M. Inglis. 1986. Evidence that the “active center” of the herpes simplex virus thymidine kinase involves an interaction between three distinct regions of the polypeptide. J. Gen. Virol. 67:753-758. [DOI] [PubMed] [Google Scholar]
- 8.Efstathiou, S., S. Kemp, G. Darby, and A. C. Minson. 1989. The role of herpes simplex virus type 1 thymidine kinase in pathogenesis. J. Gen. Virol. 70:869-879. [DOI] [PubMed] [Google Scholar]
- 9.Englund, J. A., M. E. Zimmerman, E. M. Swierkosz, J. L. Goodman, D. R. Scholl, and H. H. Balfour. 1990. Herpes simplex virus resistant to acyclovir: a study in a tertiary care center. Ann. Intern. Med. 112:416-422. [DOI] [PubMed] [Google Scholar]
- 10.Evans, J. S., K. P. Lock, B. A. Levine, J. N. Champness, M. R. Sanderson, W. C. Summers, P. J. McLeish, and A. Buchan. 1998. Herpes thymidine kinases: laxity and resistance by design. J. Gen. Virol. 79:2093-2092. [DOI] [PubMed] [Google Scholar]
- 11.Gaudreau, A., E. Hill, H. H. Balfour, A. Erice, and G. Boivin. 1998. Phenotypic and genotypic characterization of acyclovir-resistant herpes simplex viruses from immunocompromised patients. J. Infect. Dis. 178:297-303. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert, C., J. Bestman-Smith, and G. Boivin. 2002. Resistance of herpes viruses to antiviral drugs: clinical impacts and molecular mechanisms. Drug Resist. Update 5:88-114. [DOI] [PubMed] [Google Scholar]
- 13.Higuchi, R. 1990. Recombinant PCR, p. 177-183. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press Inc., San Diego, Calif.
- 14.Kudo, E., H. Shiota, T. Naito, K. Satake, and M. Itakura. 1998. Polymorphisms of thymidine kinase gene in herpes simplex virus type 1: analysis of clinical isolates from herpetic keratitis patients and laboratory strains. J. Med. Virol. 56:151-158. [DOI] [PubMed] [Google Scholar]
- 15.Larder, B. A., Y. C. Cheng, and G. Darby. 1983. Characterization of abnormal thymidine kinases induced by drug-resistant strains of herpes simplex virus type1. J. Gen. Virol. 64:523-532. [DOI] [PubMed] [Google Scholar]
- 16.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1989. Strategies for cloning in plasmid vectors, p. 1.53-1.73. In C. Nolan (ed.), Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Morfin, F., G. Souillet, K. Bilger, T. Ooka, M. Aymard, and D. Thouvenot. 2000. Genetic characterization of thymidine kinase from acyclovir-resistant and -susceptible herpes simplex virus type 1 isolated from bone marrow transplant recipients. J. Infect. Dis. 182:290-293. [DOI] [PubMed] [Google Scholar]
- 18.Morfin, F., D. Thouvenot, M. De Turenne-Tessier, B. Lina, M. Aymard, and T. Ooka. 1999. Phenotypic and genetic characterization of thymidine kinase from clinical strains of varicella-zoster virus resistant to acyclovir. Antimicrob. Agents Chemother. 43:2412-2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peterson, A., K. Grobe, B. Lindner, M. Schlaak, and W. M. Becker. 1997. Comparison of natural and recombinant isoforms of grass pollen allergens. Electrophoresis 18:819-825. [DOI] [PubMed] [Google Scholar]
- 20.Sasadeusz, J. J., F. Tufaro, S. Safrin, K. Schubert, M. M. Hubinette, P. K. Cheung, and S. L. Sacks. 1997. Homopolymer mutational hot spots mediate herpes simplex virus resistance to acyclovir. J. Virol. 71:3872-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmit, I., and G. Boivin. 1999. Characterization of the DNA polymerase and thymidine kinase genes of herpes simplex virus isolates from AIDS patients in whom acyclovir and foscarnet therapy sequentially failed. J. Infect. Dis. 180:487-490. [DOI] [PubMed] [Google Scholar]
- 22.Suzutani, T., M. Saijo, M. Nagamine, M. Ogasawara, and M. Azuma. 2000. Rapid phenotypic characterization method for herpes simplex virus and varicella-zoster virus thymidine kinases to screen for acyclovir-resistant viral infection. J. Clin. Microbiol. 38:1839-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Biotechnol. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]