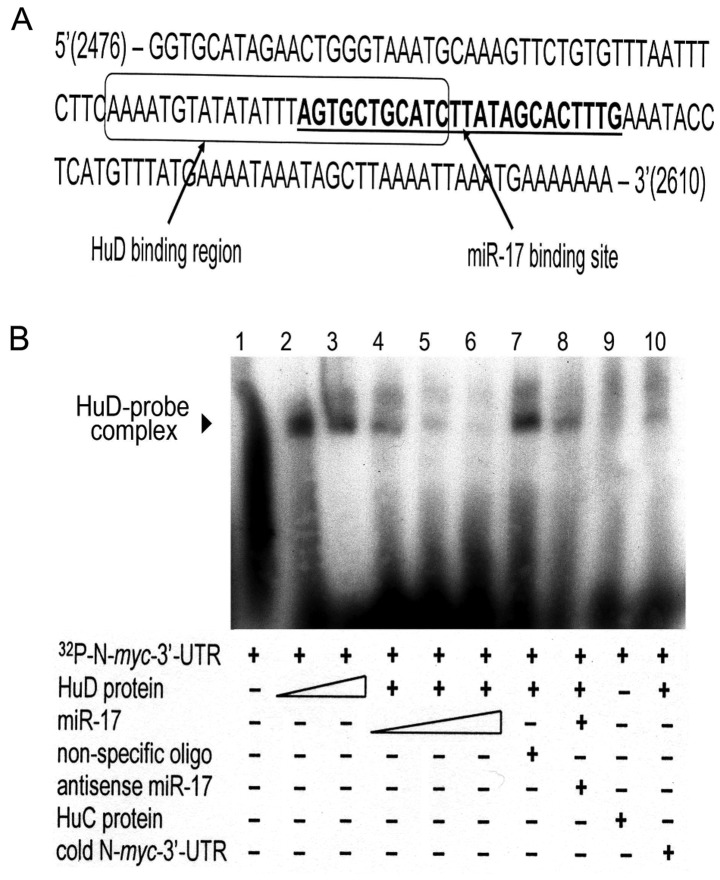
Figure 4.
HuD competes with miR-17 for an overlapping binding site in the N-myc 3′-UTR. (A) Sequence analysis revealed that HuD and miR-17 bind to adjacent regions in the N-myc 3′-UTR. The box indicates the AU-rich binding domain of HuD; the miR-17 region is indicated in underlined boldface type. Eleven bases are common to both. (B) A gel mobility shift assay (GMSA) revealed formation of the N-myc-HuD protein complex (lanes 2 and 3) and a concentration-dependent decrease in formation of the complex with increasing amounts of miR-17 (lanes 4–6). The complex amount was unaltered when non-specific oligos were substituted for miR-17 (lane 7), present but slightly decreased when miR-17 was pre-annealed with its antisense (lane 8), absent when HuC was substituted for HuD in the reaction (lane 9), and barely detectable when the labeled probe was diluted with saturating amounts of unlabeled probe (lane 10).