Figure 3.
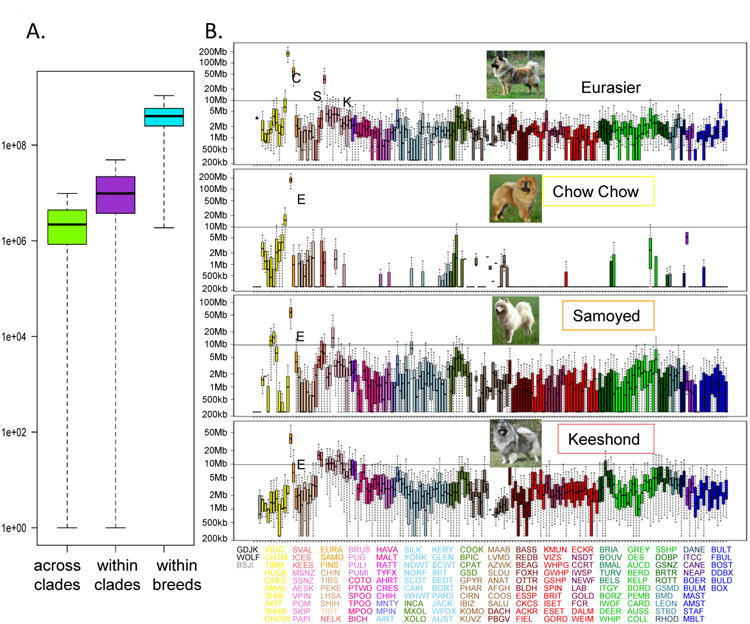
Gross haplotype sharing across breeds. A) Boxplot of total haplotype sharing between all pairs of dogs from breeds within the same clade, across different clades and within the same breed. The difference between the distributions is highly significant, p<2e-16. B) Example of haplotype sharing between three breeds (Samoyed, Chow Chow and Keeshond) and a fourth (Eurasier) that was created as a composite of the other three. Combined haplotype length is displayed on the y-axis, 169 breeds and populations are listed on the x-axis in the order they appear on the cladogram starting with the jackal and continuing counter-clockwise. Haplotype sharing of zero is set at 250,000 for graphing, a value just below what is detected in this analysis. Breeds are colored by clade. 95% significance level is indicated by the horizontal line. Breed abbreviations are listed under the graph, in the order they appear and colored by clade.
