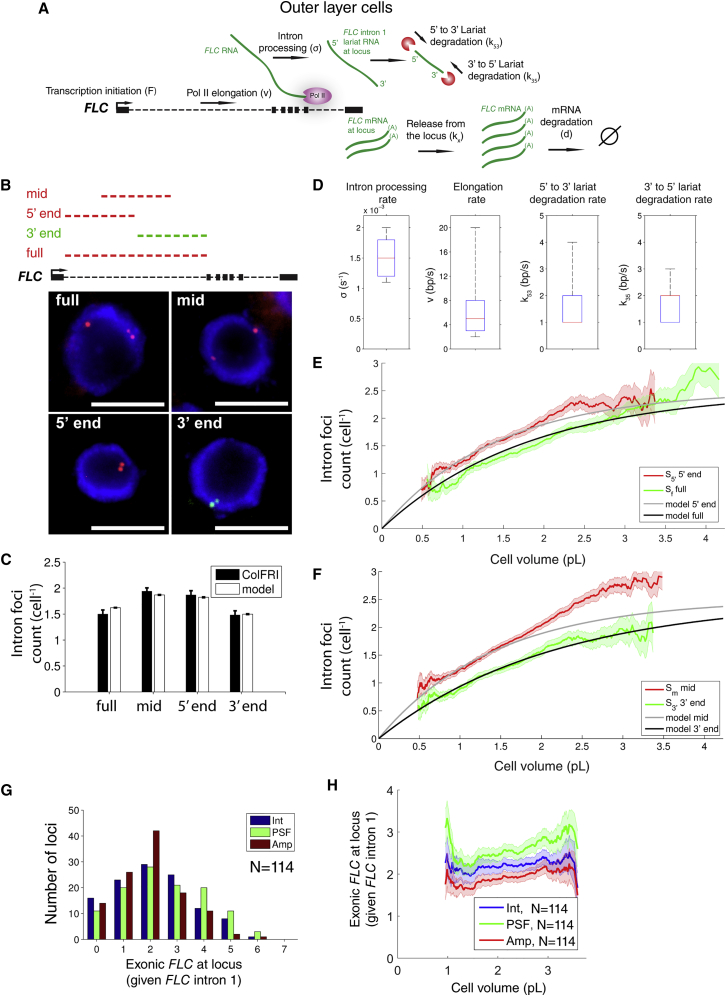
Figure 3.
Quantification of Pol II Elongation, FLC Intron 1 Processing, Lariat Degradation, and mRNA Release from the Locus
(A) Schematic of different processes contributing to FLC intron 1 life cycle: transcription initiation rate (F), Pol II elongation (v), and intron processing rate (σ). Time scale σ−1 indicates the time interval between completion of intron 1 transcription and start of lariat degradation, which can occur from 5′ to 3′ end (rate: k53) and/or 3′ to 5′ end (rate: k35). Also shown is mRNA release from the locus (rate: kx) and subsequent degradation (rate: d).
(B) Fluorescence localization (z-stack projection) of four different FLC intron 1 probe sets, as indicated on schematic: full-length (full, red), 5′ end (red), middle (mid, red), and 3′ end (green). All images are overlays of respective intron 1 signal and DAPI stain (blue) in representative outer layer root cells. Scale bar, 5 μm.
(C) Average foci counts per cell for the four probe sets described in (B) from outer layer cells together with analytical model fits (mean and SEM using all allowed parameter values, see text and STAR Methods). Number of cells analyzed, respectively, for full, mid, 5′, and 3′: 200, 382, 326, and 326 pooled from three, four, four, and four biological replicates, respectively. Error bars denote SEM.
(D) Marginal distributions for intron processing (σ), elongation (v), 5′ to 3′ lariat degradation (k53), and 3′ to 5′ lariat degradation (k35) rates that generate good fits to the data shown in (C) according to a χ2 test (degrees of freedom k = 4, acceptance probability p ≥ 0.1) with the “missing” length fraction 1/3. Box plots indicate minimum, 25% quantile, median, 75% quantile, and maximal values.
(E) Volume dependence of average cellular foci counts per cell in outer layer cells for FLC intron 1: full-length (full) and 5′ end. Number of cells analyzed as in (C). Also shown are analytical model fits (black, gray) with parameter values that generated good fits to population averages in (C): v = 3 bp/s, σ = 1.5 × 10−3 s−1, k53 = 2 bp/s, and k35 = 2 bp/s. Error lines denote SEM as function of volume.
(F) As in (E) but for middle (mid) and 3′ end FLC intron 1 probe sets.
(G) Locus-associated exonic FLC RNA (Rloc) distributions given presence of full-length FLC intron 1 signal from outer layer cells. Three different quantification methods were used on the same experimental dataset (n = 114 cells pooled from four biological replicates): integration of 2D Gaussian fit to the spatial intensity profile (Int), superposition of point spread functions (PSF), and superposition of amplitudes of PSF at the locus (Amp) (STAR Methods; Mueller et al., 2013).
(H) Volume dependence of average locus-associated exonic FLC RNA levels. Data and three different quantification methods as described in (G). Error lines denote SEM as a function of volume.