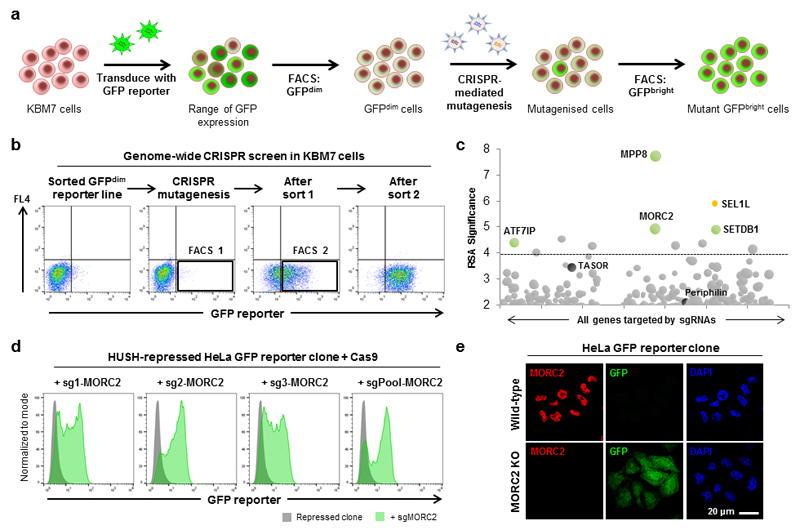
Figure 1. A genome-wide CRISPR/Cas9-mediated forward genetic screen identifies an essential role for MORC2 in transgene silencing by the HUSH complex.
(a,b) A genome-wide CRISPR screen to identify genes required for transgene silencing. Cas9 was expressed in a population of GFPdim KBM7 cells harboring epigenetically repressed transgenes, and genome-wide mutagenesis carried out using the GeCKO v2 sgRNA library (a). Mutant GFPbright cells containing gene disruption events that prevented reporter repression were isolated through two sequential rounds of FACS (b). Black boxes indicate approximate sorting gates. (c) Bubble plot illustrating the hits from the screen. All genes targeted by sgRNAs are arranged alphabetically by gene name on the x-axis, with their statistical significance as determined by the RSA algorithm plotted on the y-axis. Bubble size is proportional to the number of active sgRNAs for each gene. Colored bubbles represent validated hits; the HUSH complex subunits TASOR and Periphilin (black) did not reach statistical significance, while SEL1L (orange) is involved in the degradation of the GFP fusion protein14. A fully annotated plot is provided in Fig. S1. (d,e) MORC2 is required for transgene silencing by the HUSH complex in HeLa cells. CRISPR/Cas9-mediated disruption of MORC2 results in derepression of a HUSH-repressed reporter as measured by flow cytometry (d) or immunofluorescence microscopy (e).