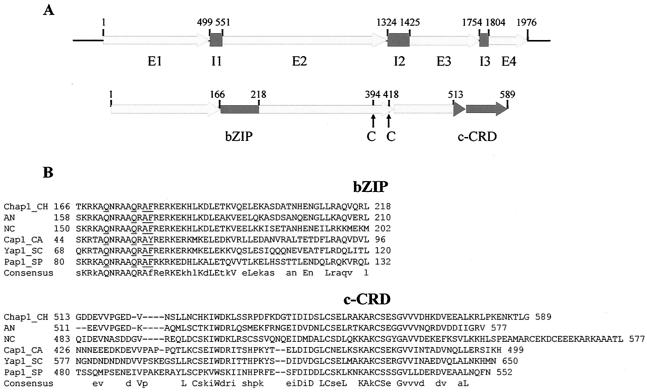
FIG. 1.
Structural organization of CHAP1. A. Upper map, organization of the gene: E1 to E4 indicate exons; I1 to I3 indicate introns. Lower map, predicted protein: bZIP, basic leucine zipper domain; c-CRD, C-terminal cysteine-rich domain; arrows indicate cysteine residues C394 and C418 belonging to the n-CRD (N-terminal cysteine-rich domain). For full details of the gene structure and domains, see GenBank accession no. AY486156. B. Alignment of bZIP and c-CRD domains of the YAP proteins. Consensus: uppercase letters indicate identity, lowercase letters indicate 50% or more similar residues. Residues characteristic of the YAP protein family are underlined. Abbreviations: Ch (CHAP1), Cochliobolus heterostrophus; An (AP-1-like protein, EAA62093), Aspergillus nidulans; Nc (AP-1-like protein, CAB91681), Neurospora crassa; Sp (Pap1, NP_593662), Schizosaccharomyces pombe; Ca (Cap1, EAL02784), Candida albicans; Sc (Yap1, NP_013707), Saccharomyces cerevisiae.