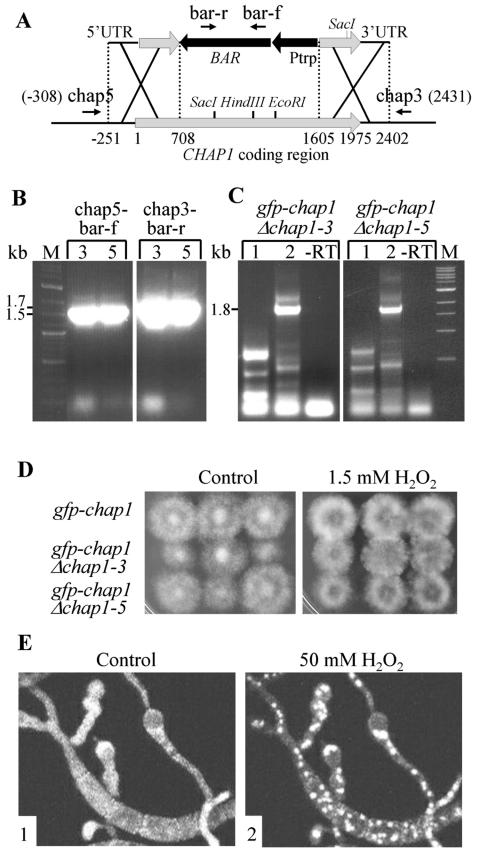
FIG. 8.
A GFP-CHAP1 fusion protein is functional in the absence of the resident CHAP1 copy. A. Deletion of the wild-type copy of CHAP1 in a GFP-CHAP1 reporter strain. BAR, phosphinothricin acetyltransferase gene; Ptrp, constitutive promoter; bar-r, bar-f, chap5, and chap3 are the primers used to confirm double-crossover integration. B. PCR verification of double-crossover integration at the CHAP1 locus in two transformants, 3 (each left lane) and 5 (each right lane). The primer pairs used for PCR were chap5 and bar-f and chap3 and bar-r. These primer pairs did not yield any product in reactions with template DNA from the wild type or the parent GFP-CHAP1 strain. Predicted product sizes are indicated at the left of the gel photo. C. Reverse transcription-PCR detection of GFP-CHAP1 but not native CHAP1 transcripts in transformants 3 and 5. Lanes 1 and 2 show reaction products with primer pairs chap-m and chap-s, and gfp-f and chap-s, respectively (described in Fig. 6A). Primer chap-m crosses the junction between genomic sequence and gfp and therefore can give a product only if the wild-type transcript is present (Fig. 6). Control reactions without reverse transcriptase (−RT) were done with primers gfp-f and chap-s. The 1.8-kb band present in lanes 2 is of the predicted size and corresponds to the transcript encoding the fusion protein (as in Fig. 6B). The lower-molecular-weight bands are nonspecific products. D. Complementation of the hydrogen peroxide-sensitive phenotype by the GFP-CHAP1 fusion protein. Left panel, control plate; right panel, 1.5 mM H2O2. The genotypes of the three strains tested are indicated at left. All strains grew equally well on both media. E. Nuclear localization of GFP-CHAP1 upon exposure to 50 mM H2O2 in the absence of native CHAP1. Panels: 1, conidia and germ tubes growing on a glass slide prior to hydrogen peroxide treatment; 2, 5 min after addition of 50 mM H2O2.