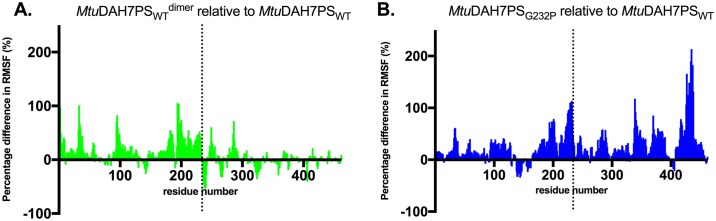
Fig 6. Percentage difference in residue RMSF values.
Percentage difference in residue RMSF values between (A) tetrameric MtuDAH7PSWT and dimeric variant MtuDAH7PSG232P and (B) tetrameric MtuDAH7PSWT and theoretical dimer MtuDAH7PSwtdimer. The dashed line indicates the position of the substitution G232P along the amino acid sequence of MtuDAH7PS. Residue RMSF values were measured in reference to the corresponding MD average conformation using trajectories obtained from equilibrated time period in the MD simulation for each system (50–464.1 ns, 70–429.9 ns and 120–540.9 ns for MtuDAH7PSG232P, MtuDAH7PSWT and theoretical dimer MtuDAH7PSwtdimer, respectively). Chain-average RMSF values were calculated and compared. Percentage difference is calculated by (RMSFdimer − RMSFtetramer) ÷ RMSFtetramer × 100.