Figure 6. MYRF-1(G274R) mutant interferes with MYRF’s normal function.
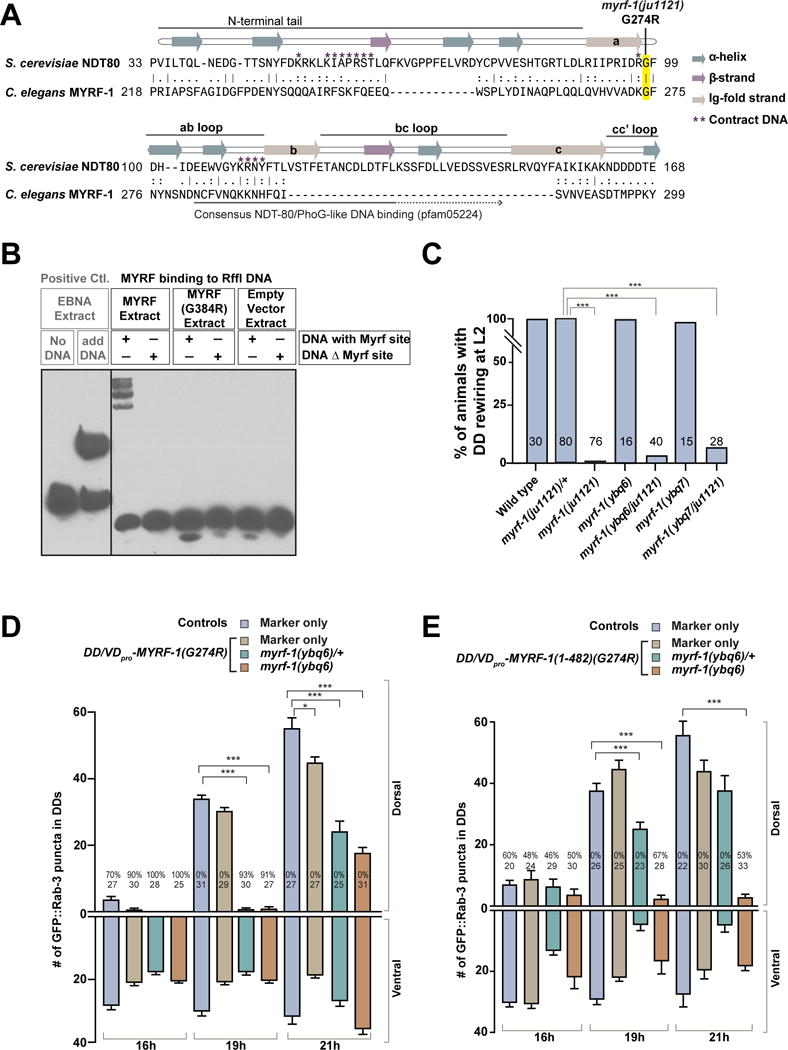
A. Alignment between segments of DNA-binding domains of MYRF-1 and yeast Ndt80. Annotation of Ndt80 strands are redrawn based on Lamoureux et al. 2002.
B. Electrophoretic mobility shift assay (EMSA) testing mouse MYRF binding on Rffl DNA. MYRF(R384R) mutation is equivalent to MYRF-1(G274R) in C. elegans.
C. Percentage of animals with normal DD rewiring. Animals of myrf-1(ybq6/ju1121) are cross progenies of myrf-1(ybq6)/mIn1 and myrf-1(ju1121)/mIn1. myrf-1(ybq7/ju1121) was generated similarly. Pearson’s chi-squared test (***P<0.001).
D. Number of synapses in animals expressing unc-25pro-MYRF-1(G274R) (ybqIs27) labeled by flp-13pro-GFP∷RAB-3 (ybqIs47) is shown as mean ± SEM; t-test (*P<0.05, ***P<0.001); the number of animals analyzed is shown on each bar; %, penetrance for animals with no dorsal synapse.
E. Number of synapses labeled by flp-13pro-GFP∷RAB-3 (ybqIs47) in animals expressing unc-25pro-N-MYRF-1(G274R) (ybqEx517, ybqEx541) is shown as mean ± SEM; t-test (***P<0.001); the number of animals analyzed is shown on each bar; %, penetrance for animals with no dorsal synapse.
(See also Figure S7.)
