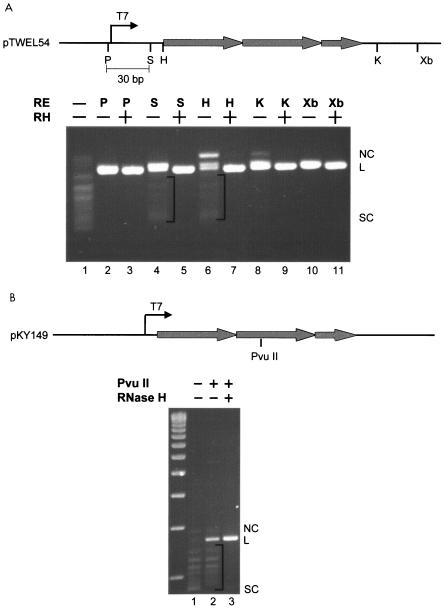
FIG. 3.
Extent of R-loop formation on a 2.5-Sγ3 repeat segment. (A) The top panel is a diagram representing a part of pTW-EL54. Shaded thick arrows represent Sγ3 repeats. The lower panel shows the digestion products of T7 RNA polymerase-transcribed pTW-EL54 (R-loop). Irrelevant lanes between lanes 7 and 8 have been removed. The location of the R-loop was probed by testing for the cleavability of restriction sites upstream and downstream of the switch region. Bands below the linear position represent uncut plasmid due to the R-loop formation. RNase H removes the RNA of the R-loop, thereby permitting the two DNA strands to anneal and become cleavable by the restriction enzymes. The brackets indicate the undigested plasmid due to R-loop formation in the region of the restriction enzyme sites. (B) R-loop status of the middle of the 125-bp switch region. A PvuII restriction site within the middle of the 2.5-repeat switch region was tested for cleavability. NC, nicked circular; L, linear; SC, supercoiled; T7, T7 promoter; P, PstI; S, SacI; H, HindIII; K, KpnI; X, XbaI.