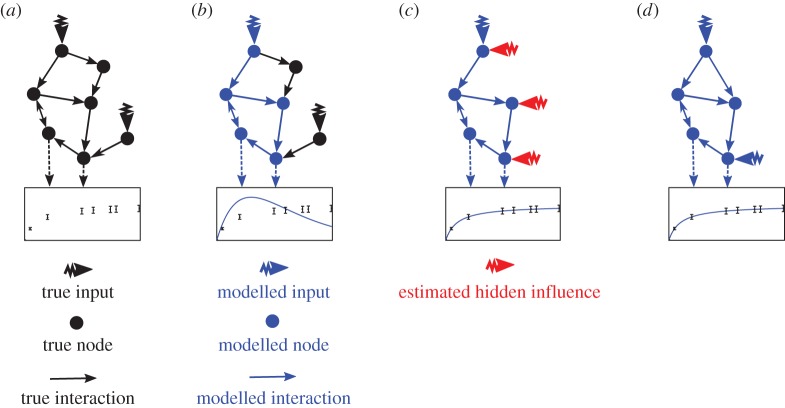
Figure 1.
Illustration of the Bayesian approach to estimating hidden influence variables in ODE-based models. Here, a fictional interaction network is shown. (a) True system (black) with two inputs and one fictive observable measurement as a combination of two nodes (box). (b) In reality, the available knowledge represented by an a priori model (blue) does not necessarily cover the whole system but only a part of the true system. Hence the nominal model leads to an unsatisfactory fit with the observable measurement. This may caused by exogenous influences or by misspecified molecular interactions (i.e. missing or wrong edges in the interaction network). (c) Our approach aims for estimating these hidden influences (red) and the directly involved molecular species. (d) Some of the estimated hidden influences may correspond to missing or wrong molecular interactions within the system. Hence, in a last step, our method tries to further distinguish between intrinsic and exogenous hidden influences. We therefore identify erroneous parts of the nominal ODE system and give detailed hints for their correction.