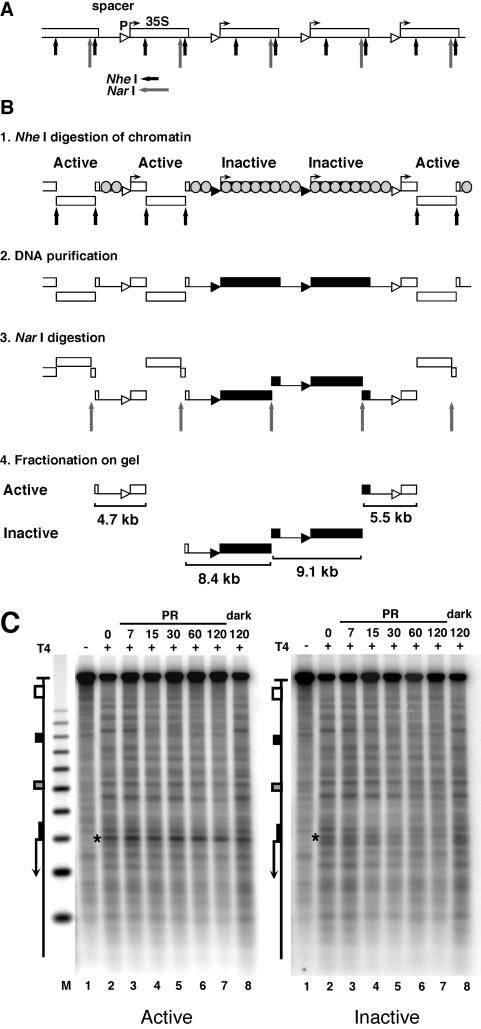
FIG. 3.
Inhibition of photoreactivation in active promoters. (A) Schematic illustration of five rRNA gene repeats with the 35S rRNA genes (boxes), promoters (triangles), and spacer and restriction sites for NheI (short arrows) and NarI (long arrows). (B) Fractionation procedure for active and inactive 35S promoters in rRNA gene chromatin. White and black triangles and boxes depict active and inactive promoters and genes, respectively. Spacers and inactive genes are packaged in nucleosomes (grey-shaded circles); active genes are free of nucleosomes (11). Panel 1: nuclear chromatin was digested with NheI to release transcriptionally active genes (30, 32) and a fragment containing an rRNA gene spacer flanked by active genes. All other promoters remain in long, uncut chromatin fragments. Panel 2: DNA was purified. Panel 3: DNA was digested with NarI, which generates spacers containing fragments of 4.7, 8.4, 9.1, and 5.5 kb. Panel 4: the DNA was fractionated on an agarose gel, and fragments containing active promoters (open triangles; 4.7 and 5.5 kb) and inactive promoters (black triangles; 8.4 and 9.1 kb) were excised and purified. (C) AMY3cells were irradiated as described for Fig. 2. DNA fragments containing promoters of active and inactive genes were purified (as depicted in panel B), digested with NheI and NdeI, cut at CPDs with T4-endoV (lanes 2 to 8), fractionated on alkaline agarose gels, and blotted and hybridized with strand-specific probes for the bottom strand (see Fig. 1A). Lanes 1, same as lanes 2 but with no T4-endoV treatment; M, size marker as described for Fig. 2. The 35S promoter is marked with a star.