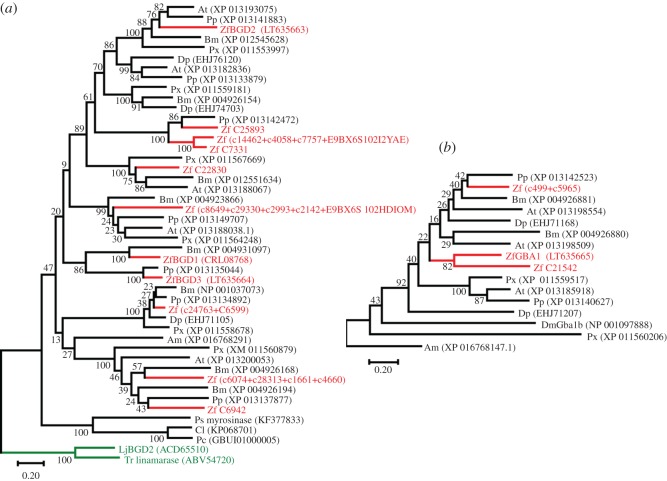
Figure 4.
Models of the evolutionary history of insect GH 1 β-glucosidases (a) and GH 30 glucocerebrosidases (b). Analyses were carried out on protein sequences in MEGA7 using the maximum-likelihood method with a JTT model, and a discrete Gamma distribution. The branch lengths are measured in the number of substitutions per site, and 1000 bootstrap replicates were carried out. Am, Apis mellifera; At, Amyelois transitella; Bm, Bombyx mori; Cl, Chrysomela lapponica; Dm, Drosophila melanogaster; Dp, Danaus plexippus; LJ, Lotus japonicus (green); Pc, Phaedon cochleariae; Pp, Papilio polytes; Px, Plutella xylostella; Ps, Phyllotreta striolata; Tr, Trifolium repens (green); Zf, Zygaena filipendulae (red). (GenBank accession numbers shown).