Figure 2.
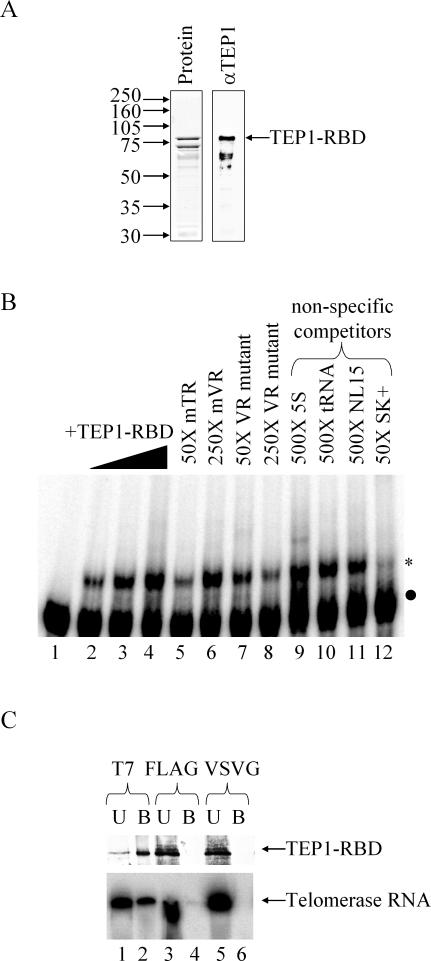
Partially purified murine TEP1–RBD interacts with TR in vitro but has limited specificity. (A) Amino acids 201–871 of murine TEP1 were expressed in E.coli and partially purified using the hexahistidine tag derived from the pET28a vector. Shown are a Coomassie-stained gel (left panel) and western blot (right panel) using anti-TEP1 polyclonal antibodies. (B) EMSA of nt 1–223 of mouse TR incubated with TEP1–RBD. Five fmol 32P-labeled probe was incubated with 0, 5, 20 and 80 ng TEP1–RBD (lanes 1–4). Probe was competed off with 50× full-length TR (lane 5) and increasing amounts of mVR double point mutant (lanes 7 and 8), but not wild-type mVR (lane 6). The 250× unlabeled 5S Ribosomal RNA, tRNAphe or an artificial RNA (NL15) do not compete with the probe (lanes 7–9), but 50× excess of an artificial RNA derived from the pBluescript polylinker region does compete efficiently (lane 10). Labeled RNA is indicated with a black dot and shifted complexes are indicated with an asterisk. (C) Immunoprecipitation of TEP1–RBD from binding reactions using anti-T7 monoclonal antibody co-immunoprecipitates the TR transcript. Equivalent amounts of the bound (B) and unbound (U) fraction were analyzed by either western blot using anti-TEP1 polyclonal antibody (upper panel) or by fractionation on a 10% acrylamide/8 M urea gel (lower panel). The latter gel was dried and radioactive bands visualized by phosphorimager analysis. Antibodies to the T7 epitope (lanes 1 and 2), but not antibodies to the FLAG (lanes 3 and 4) and VSVG (lanes 5 and 6) epitopes, immunoprecipitated both TEP1–RBD and TR.