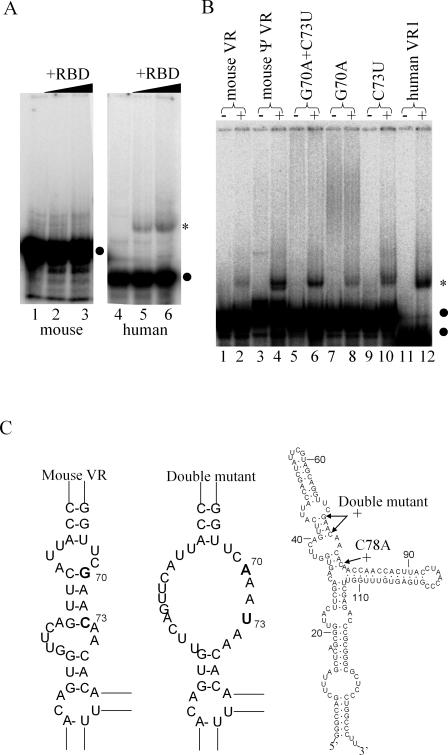
Figure 3.
Mutagenesis of murine VR enhances its binding to TEP1–RBD. (A) EMSA of VR–TEP1–RBD complexes. Binding reactions contained 5 fmol of 32P-labeled murine VR (left panel) or human VR1 (right panel) incubated with 0 (lanes 1 and 4), 20 (lanes 2 and 5) and 80 (lanes 3 and 6) ng of TEP1–RBD. (B) EMSA of VR probes (5 fmol) without (−) or with (+) 80 ng TEP1–RBD in the binding reaction. Wild-type mouse VR interacts poorly or not at all with TEP1–RBD (lanes 1 and 2), but a highly related sequence in the mouse genome that is not expressed binds more strongly (lanes 3 and 4). A VR double point mutant G70A, C73U (lanes 5 and 6) binds more strongly to TEP1–RBD. Also shown are the corresponding single mutants in mouse VR (lanes 7–10) and human VR1 (lanes 11 and 12). Labeled RNAs are indicated with a black dot and shifted complexes are indicated with an asterisk. (C) The thermodynamically predicted secondary structure of the wild-type (left structure) and double point mutant (center structure) mouse VR, focusing on the regions of interest only. Also shown is the entire predicted structure of mouse VR, along with the position of the double point mutant and the C78 to A mutation, which has enhanced binding.