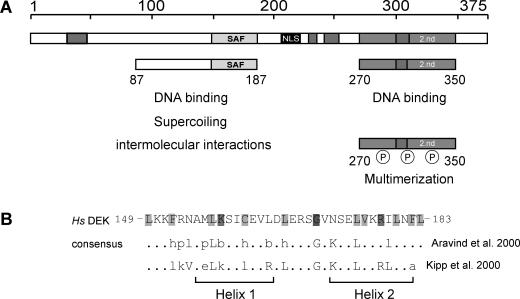
Figure 1.
The human DEK protein—an overview. (A) Schematic representation of DEK with the known functional domains. The central SAF or SAP box (149–183) is shown in light gray, the nuclear localization sequence (NLS) in black, the four highly acidic regions in dark gray and the second DNA-binding domain, located at the C-terminal region, is represented by bright gray (2.nd). The 87 to 187 peptide mediates DNA binding, induces supercoiling in relaxed circular DNA and promotes the formation of large nucleoprotein complexes (‘intermolecular interactions’) (14). The region from residues 270 to 350 harbors most of the mapped phosphorylation sites. When phosphorylated, the overall DEK–DNA interaction is reduced and DEK–DEK interactions are promoted (‘multimerization’) (11,14,15). (B) Primary structure of the SAF or SAP box of human DEK in comparison with consensus sequences derived by Kipp et al. (17) and Aravind and Koonin (16) and a schematic representation of the predicted secondary structure (two amphipathic helices separated by a region with an invariant glycine). Light gray, small and hydrophobic residues (A, V, L, I, M, F, W); bright gray, charged residues (D, E, R, K); dark gray, glycin (modified from: http://www.sanger.ac.uk/Software/Pfam, search term: SAP). Abbreviations used: h (hydrophobic) and l (aliphatic): Y, F, W, L, I, V, M, A; p (polar): S, T, Q, N, E, D, R, K, H; and b (bulky): K, R, E, Q, W, F, Y, L, M, I; Hs, Homo sapiens. Amino acids indicated in capitals contribute to the consensus sequence in more then 90% of all investigated SAP box proteins (16).