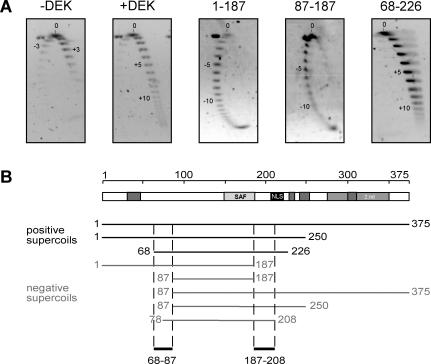
Figure 5.
Changing the topology of relaxed circular DNA. (A) The 2D gel electrophoresis. A standard topology assay was performed without protein (−DEK) or with recombinant full-length his-DEK (+DEK), 1–178, 87–187 or 68–226 at molar ratios of 150 (DEK/DNA). The purified DNA was separated by standard agarose gel electrophoresis for the first dimension (16 h, 2 V/cm) (from top to bottom). The gel was rotated by 90°, incubated in 0.25 μg/μl chloroquin (in 0.5× TBE) followed by a run for the second dimension (3 h, 4 V/cm) (from left to right). DNA was visualized by SybrGold staining. Direction (−; +) and number of introduces supercoils are indicated in the figures. (B) Schematic overview of all DEK-derived peptides tested in 2D gel electrophoresis. The direction of introduced supercoils is indicated for each peptide. Sequences 68–87 and 187–208 are necessary for the introduction of positive supercoils.