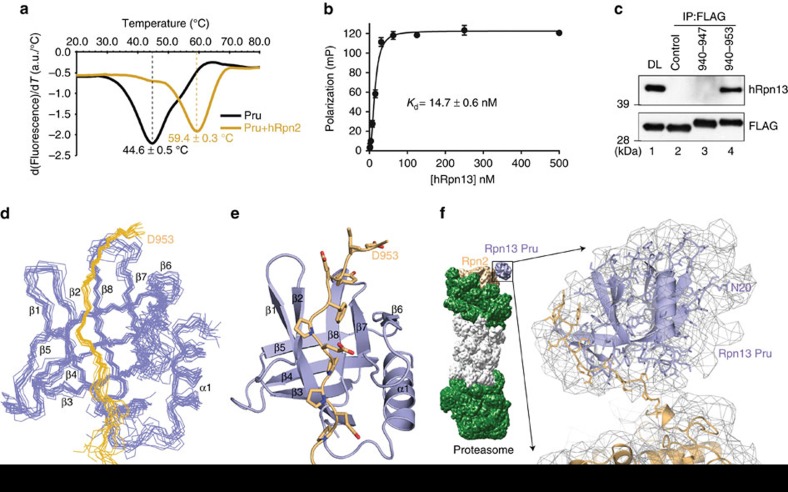
Figure 1. Structure of hRpn13 at the proteasome.
(a) hRpn13 Pru alone (black) or with hRpn2 (940–953) (orange) was heated from 20 to 85 °C at a rate of 1 °C per min. The melting temperature was calculated from the first derivative of tryptophan emission intensities at 350 nm (a.u., arbitrary unit). (b) 10 nM FITC-labelled hRpn2 (940–953) was incubated with varying concentrations of full-length hRpn13 as indicated in triplicate to measure a binding constant by FP. Corresponding values for the probe alone were subtracted from measurements of the complex and the final numbers plotted against hRpn13 concentration and fit by nonlinear regression to a Hill slope model. The error bar represents the s.d. of each data point to the average value. (c) Cell lysates or immunoprecipitates derived by anti-FLAG antibodies from HCT116 cells expressing FLAG-EGFP (control), FLAG-EGFP-hRpn2 (940–947) or FLAG-EGFP-hRpn2 (940–953) were subjected to immunoprobing, as indicated. Direct loading (DL) of the lysates from FLAG-EGFP-expressing HCT116 cells is also included. (d) Backbone heavy atoms for the 12 lowest energy structures with best geometry for the hRpn13 Pru-hRpn2 (940–953) complex with hRpn13 displayed in periwinkle blue and hRpn2 in light orange. (e) Ribbon diagram for the hRpn13 Pru-hRpn2 (940–953) structure depicting the classic pleckstrin homology fold of hRpn13 Pru (periwinkle blue) with the hRpn2 peptide (light orange) extended across a β-strand surface. hRpn2 nitrogen and oxygen atoms are displayed in blue and red, respectively. (f) The hRpn13 Pru-hRpn2 (940–953) structure is modelled into a cryoEM reconstruction (EMD-2594, displayed in grey) from S. cerevisiae 26S proteasome50 that includes the scRpn2 PC repeat region (PDB code 4CR2). The N-terminal 19 amino acids of hRpn13 Pru are randomly coiled and most likely contribute to the extra density displayed near residue N20.