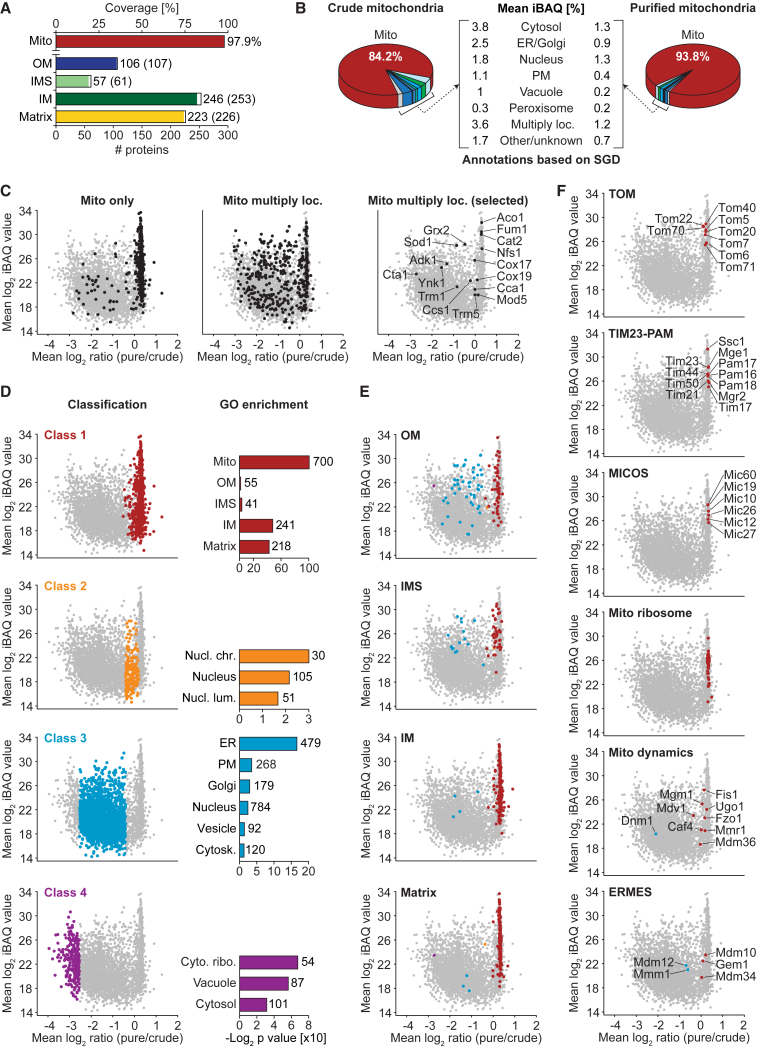
Figure 2.
Quantitative Deep Proteome Analysis of Yeast Mitochondria by SILAC-MS
(A) Coverage of proteins of distinct submitochondrial localization. Proteins identified in pure/crude mitochondria were categorized as outer membrane (OM), intermembrane space (IMS), inner membrane (IM), or matrix protein based on GO annotations. Shown is the number of proteins assigned to a given term and (in parentheses) the total number of proteins with this annotation. Mito, sum of all proteins in OM, IMS, IM, and matrix.
(B) Protein composition of crude versus gradient-purified mitochondria. Shown are mean percentages of intensity-based absolute quantification (iBAQ) values (n = 4). Loc., localized; PM, plasma membrane; SGD, Saccharomyces Genome Database.
(C) Ratio-intensity plots of quantified proteins (n = 4). Proteins exclusively localized to mitochondria (left), to mitochondria and other organelles (middle), and selected proteins with multiple subcellular localization (right) are highlighted.
(D) Distribution of proteins grouped into distinct classes by statistical data analysis (left panel) and selected GO cellular component terms significantly enriched in each class (right panel) with the number of proteins assigned. Cyto. ribo., cytosolic ribosome; Cytosk., cytoskeleton; Nucl. chr./lum., nuclear chromosome/lumen.
(E and F) Ratio-intensity plots as shown in (D) highlighting mitochondrial subcompartment annotations (E) and subunits of the TOM, TIM23-PAM, MICOS, and ERMES complex as well as mitochondrial ribosomal proteins and proteins involved in mitochondrial dynamics (F). Coloring reflects the class a protein was assigned to.