Figure 5.
Profiling of Suborganellar Localization and Membrane Topology of Mitochondrial Proteins
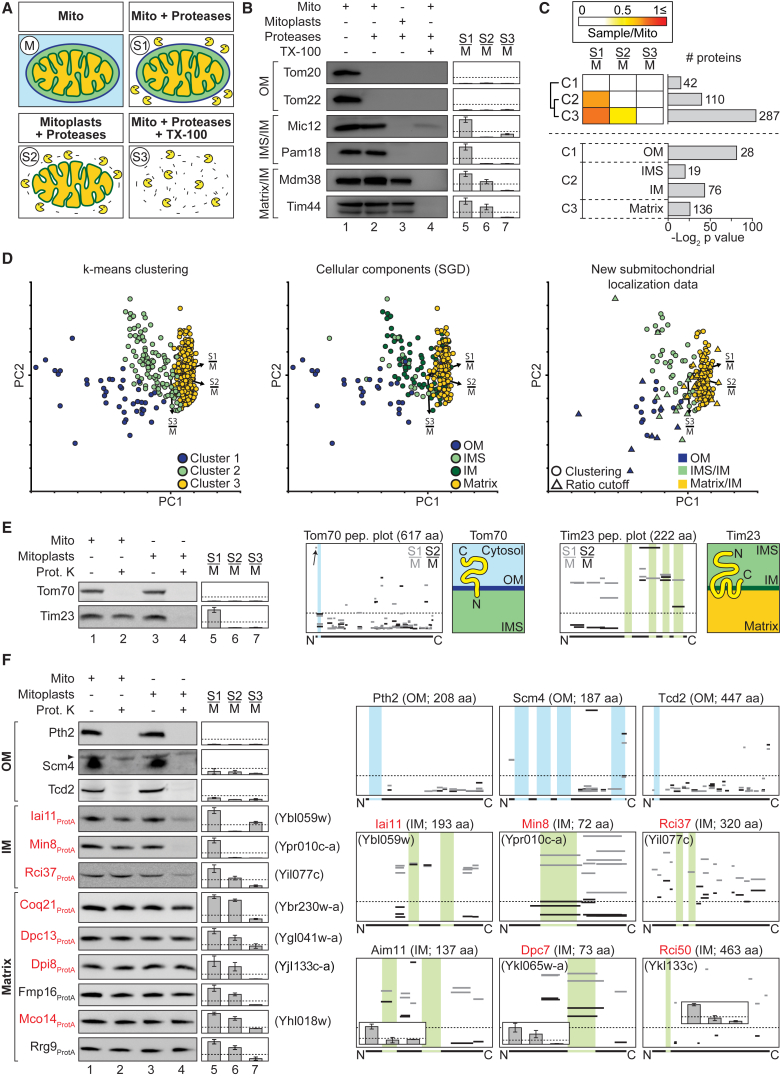
(A and B) Protease accessibility assay. Gradient-purified mitochondria (M, S1), mitoplasts (S2), and mitochondrial Triton X-100 (TX-100) lysates (S3) were treated with proteases (proteinase K and trypsin) as indicated. (B) Samples were analyzed by SDS-PAGE and immunoblotting using antisera against marker proteins of the mitochondrial outer membrane (OM), and intermembrane space (IMS)- and matrix-exposed proteins (lanes 1–4). For submitochondrial profiling, SILAC-labeled untreated mitochondria (M) and S1, S2, or S3 were mixed and analyzed by MS (n = 3) (lanes 5–7). Y axes, mean S/M protein ratios; error bars indicate SEM for n = 3 and the range for n = 2; dotted lines, S/M ratios of 0.25. IM, inner membrane.
(C) Top, k-means clustering performed based on S/M ratios of proteins (n = 3) with a ratio ≤1.1 that were present in class 1 in pure/crude experiments and showed decreasing ratios with increasing accessibility of the proteases (i.e., S1/M ≥ S2/M > S3/M). Bottom, selected GO terms significantly enriched in each cluster.
(D) Principal-component (PC) analysis of log2-transformed mean S/M ratios of the proteins present in the clusters depicted in (C) and further proteins meeting the criteria for signature plots. Shown are all proteins of C1–C3 (left), proteins with previous GO annotations (middle), and proteins without previous GO annotation that were assigned to mitochondrial subcompartments based on our experimental data (right). PC 1–3 account for 45.7%, 35.3%, and 19.0% of the variability in the data. Arrows point to the direction of increasing values for the different experimental conditions.
(E and F) Mitochondrial sublocalization and membrane protein topology. Intact mitochondria (lanes 1 and 2) or mitoplasts (lanes 3 and 4) were treated with proteinase K (Prot. K) where indicated. Marker proteins of the mitochondrial outer (Tom70) and inner membrane (Tim23) shown in (E) belong to the fractionation of Mco8ProtA (Figure S6). Bar charts (lanes 5–7) show the corresponding S/M protein ratios. Peptide plots illustrate the topology for Tom70, Tim23 (both summarized in adjacent cartoons), and selected proteins of the high-confidence mitochondrial proteome. Plotted are S/M ratios of tryptic peptides identified in submitochondrial profiling experiments. Transmembrane segments (TMHMM prediction or according to Alder et al. [2008] for Tim23) are indicated in blue (OM) or green (IM). Y axes of bar charts and peptide plots, mean S/M ratio; dotted lines indicate S/M ratios of 0.25; error bars indicate SEM for n = 3 and the range for n = 2.