Figure 5. Circadian stress network results in time‐dependent apoptotic response of human ES cell‐derived cardiac cultures.
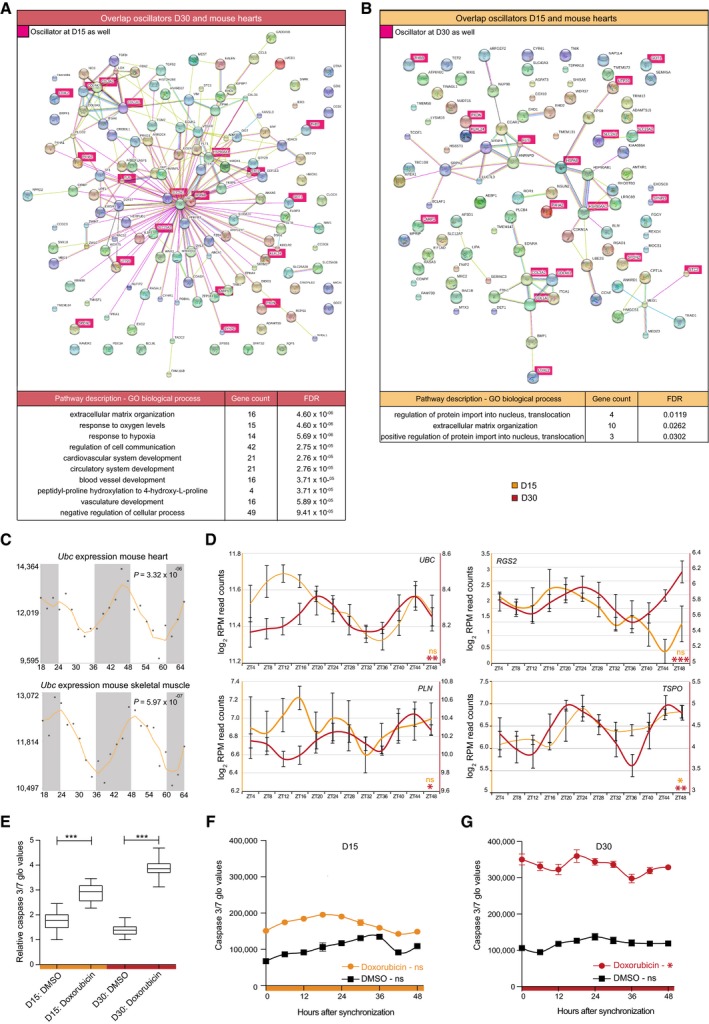
- STRING interaction network of overlapping oscillators between D30 cardiac cells and mouse hearts with corresponding GO analysis. Genes that oscillate at both D15 and D30 are depicted in pink. Mouse heart oscillators were deducted from Zhang et al 14.
- Same analysis as in (A) for D15 oscillators.
- Ubc mRNA oscillation in mouse hearts and skeletal muscle. Data were obtained from CircaDB (http://circadb.hogeneschlab.org/, deducted from 14, 83). Corresponding CircaDB P‐values are indicated.
- Expression levels of four D30 oscillatory genes (UBC, RGS2, PLN, and TSPO) across 48 h. Average log2 RPM CEL‐Seq counts of three replicates were plotted for D15 and D30 and smoothened over two time points ± s.e.m. Significance of rhythmicity across 48 h was analyzed using the JTK‐cycle algorithm (ns: not significant; * JTK P < 0.05, ** JTK P < 0.005, and *** JTK P < 0.0005).
- Apoptosis, as measured by caspase 3/7 activity, after doxorubicin administration in D15 and D30 human ES cell‐derived cardiac cells across all samples from (F) and (G). Bottom and top of the boxes are the 25th and 75th percentiles. The line within the boxes represents the median and whiskers denote the minimum and maximum values. The effect of doxorubicin versus DMSO was tested using a Mann–Whitney U‐test (***P < 0.0005).
- Apoptosis measured with 6‐h intervals in synchronized D15 cultures after administration of doxorubicin (orange) and DMSO as control (black) for 6 h. Data are represented as the mean ± s.e.m. of three independent replicates. Significance of rhythmicity across 48 h was analyzed using the RAIN algorithm and is indicated (ns: not significant).
- Same as in (F) for D30 cultures with the doxorubicin response depicted in red (RAIN, ns: not significant and *P < 0.05). Data are represented as the mean ± s.e.m. of three independent replicates.
